Abstract
Sphagneticola calendulacea is a valuable medicinal herb. With the spread of a congeneric invasive species Sphagneticola trilobata in South China, the already vulnerable S. calendulacea populations are being threatened further by natural hybridization with the invading species. In this study, we assembled and characterized the complete chloroplast genome of S. calendulacea as a resource for future studies on this species. The chloroplast genome was 151,748 bp in size, with a large single-copy (LSC) region of 83,270 bp, a small single-copy (SSC) region of 18,348 bp, separated by two inverted repeat (IR) regions of 25,065 bp each. A total of 134 genes were predicted. Phylogenetic analysis showed close relationship between S. calendulacea and Eclipta prostrata within the Heliantheae tribe.
Sphagneticola calendulacea (L.) Pruski (synonym: Wedelia chinensis) is an indigenous perennial herb in South China. This species can be found at riverbanks, coastland, and other moist habitats. With various active constituents like flavonoids, diterpenes, triterpenes, saponins, and phytosterols, S. calendulacea has been widely used in traditional medicines (Nomani et al. Citation2013). Before the introduction of the congeneric invasive species S. trilobata (Lowe et al. Citation2000), S. calendulacea was once the only Sphagneticola species in South China. In recent years, S. trilobata has rampantly spread across South China, threatening the survival of the native S. calendulacea. The threats from the invader are many-folds; besides competitive exclusion, natural hybridization with S. trilobata is also posing a great threat to S. calendulacea. Furthermore, the huge amount of pollen from the invasive species greatly reduces the chance of receiving its own pollen. With widespread hybridization, the F1 hybrid generation has recently emerged as a new competitor (Li et al. Citation2015; Wu et al. Citation2013). From our field surveys, many S. calendulacea populations have disappeared from their recorded locations. In this study, we characterized the complete chloroplast genome sequence of S. calendulacea as a resource for future studies on this species.
We sampled the fresh leaves of S. calendulacea from a coastland in Nansha, Guangzhou, China. A voucher specimen was also deposited at the Sun Yat-sen University Herbarium (SYS) with accession number STR-2017-SYS2. Total DNA was extracted using a CTAB protocol, before a library with insertion size of 500 bp was constructed and paired-end (125 bp) sequenced on an Illumina Hiseq2500 platform. Using an atpB-rbcL gene sequence (GenBank accession JQ065019.1) of S. calendulacea as seed, the chloroplast genome assembly was performed using the program NOVOPlasty (Dierckxsens et al. Citation2017) with ∼4 Gb raw reads as input. The chloroplast genome was then annotated using DOGMA (Wyman et al. Citation2004) and manually corrected.
The complete chloroplast genome sequence of S. calendulacea (GenBank accession KY828438) was 151,748 bp in length, with a large single-copy (LSC) region of 83,270 bp, a small single-copy (SSC) region of 18,348 bp, and a pair of inverted repeat (IR) regions of 25,065 bp each. The genome consisted of 134 predicted genes, including 86 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. The overall GC content was 37.48%.
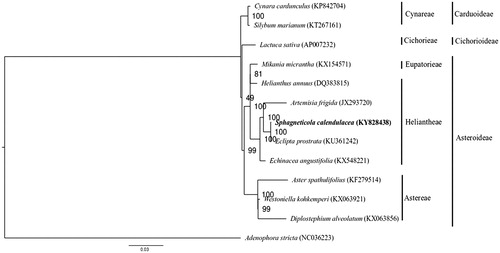
For phylogenetic analysis, the chloroplast genome sequences of Adenophora stricta (Campanulaceae) and 11 representative species of the family Asteraceae were downloaded from the NCBI GenBank database. The 86 protein-coding genes of S. calendulacea were then BLASTed against the chloroplast genome sequences of the 12 species. The corresponding orthologs were then extracted and concatenated before sequence alignment using MAFFT v7.307 (Katoh and Standley Citation2013). After determining the nucleotide-substitution model using the program jModelTest 2 (Darriba et al. Citation2012), a maximum likelihood tree was constructed with the GTR + G model implemented in the program RAxML (Stamatakis Citation2014), with A. stricta as outgroup. In the phylogenetic tree, S. calendulacea was found to be sister to Eclipta prostrata (KU361242) within the Heliantheae tribe with strong bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li HM, Ren C, Yang QE, Yuan Q. 2015. A new natural hybrid of Sphagneticola (Asteraceae, Heliantheae) from Guangdong, China. Phytotaxa. 221:71–76.
- Lowe S, Browne M, Boudjelas S, De Poorter M. 2000. 100 of the world’s worst invasive alien species. A selection from the Global Invasive Species Database. Published by the Invasive Species Specialist Group (ISSG) a Specialist Group of the Species Survival Commission (SSC) of the World Conservation Union (IUCN) pp 12.
- Nomani I, Mazumder A, Chakraborthy GS. 2013. An overview of a potent medicinal herb Wedelia chinensis (Asteraceae. Int J PharmTech Res. 5:957–964.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wu W, Zhou RC, Ni GY, Shen H, Ge XJ. 2013. Is a new invasive herb emerging? Molecular confirmation and preliminary evaluation of natural hybridization between the invasive Sphagneticola trilobata (Asteraceae) and its native congener S. calendulacea in South China. Biol Invasions. 15:75–88.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.