Abstract
Changiostyrax dolichocarpa is a critically endangered plant species occurring in central and southeastern China. Although the systematic position of Changiostyrax was still unclear, morphological characters, plastid and nSSR evidence supported that C. dolichocarpa should be separated from Sinojackia (Styracaceae). Here, we report and characterize the complete plastid genome sequence of C. dolichocarpa in an effort to provide genomic resources useful for promoting its conservation. The complete plastome is 158,821 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat (IR) regions of 26,000 bp, a large single copy (LSC) region of 88,038 bp and a small single copy (SSC) region of 18,784 bp. The plastome contains 120 genes, consisting of 83 unique protein-coding genes, 37 unique tRNA gene, and eight unique rRNA genes. The overall A/T content in the plastome of C. dolichocarpa is 62.70%. The complete plastome sequence of C. dolichocarpa will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Changiostyrax.
Changiostyrax dolichocarpa C.T.Chen (Styracaeae, formerly Sinojackia dolichocarpa) is a rare and endangered plant of the family Styracaceae (Qin et al. Citation2017) and distributed in Hubei and Hunan Province of China (Li et al. Citation2002), which also has potential horticultural value. Based on the morphological characteristics of the fruit and flower, C. dolichocarpa was recognized as a species within the independent genus (Changiostyrax) (Chen Citation1995). Although the systematic position of Changiostyrax was still unclear, morphological characters, plastid and nSSR evidence supported that C. dolichocarpa should be separated from Sinojackia (Styracaceae) (Yao et al. Citation2008). Consequently, the genetic and genomic information is urgently needed to promote its systematics research and the development of conservation value of C. dolichocarpa. Here, we report and characterize the complete plastome of C. dolichocarpa (GenBank accession number: this study). This is the first report of a complete plastome for the genus Changiostyrax.
In this study, C. dolichocarpa was sampled from Hunan Hupingshan National Nature Reserve in Hunan province of China (111.386°E, 29.591°N). A voucher specimen (H.F. Wang, sm2) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
The modified cetyltrimethyl ammonium bromide (CTAB) protocol of Doyle and Doyle (Citation1987) was used to extract genomic DNA from dry leaf tissues. The genomic DNA of each sample was quantified and analysed with Agilent 2100 BioAnalyzer. Samples yield at least 0.8 μg of DNA which were selected for subsequent libraries construction and de novo sequencing. Genomic DNA of selected samples was used to build the paired-end libraries with 200–400 bp insert size. Libraries were sequenced using BGISEQ-500 platform at BGI Shenzhen, China and produced about 8 Gb high quality per sample with 100 bp paired-end reads. Raw reads were trimmed using SOAPfilter_v2.2 with the following criteria: (1) reads with >10% base of N; (2) reads with >40% of low quality (value ≤10); (3) reads contaminated by adaptor and produced by PCR duplication. Around 6 Gb clean data were assembled against the plastome of Sinojackia rehderiana (MF179499.1) (Yu et al. Citation2017) using MITO bim v1.8 (Hahn et al. Citation2013).
The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Sinojackia xylocarpa (NC035418). The annotation was corrected with DOGMA (Wyman et al. Citation2004). A circular plastome map was generated using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The plastome of C. dolichocarpa was found to possess a total length of 158,821 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 26,000 bp, a large single copy (LSC) region of 88,038 bp and a small single copy (SSC) region of 18,784 bp. The plastome contains 120 genes, consisting of 83 unique protein-coding genes, 37 unique tRNA gene and four unique rRNA genes. Among these gene, 15 genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, rpl12, rps16, atpF, rpoC1, petB, petD, rpl16, ndhB, ndhA) possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of C. dolichocarpa is 62.70%, in which the corresponding value of the LSC, SSC, and IR region was 64.70%, 69.50%, and 57.00%, respectively.
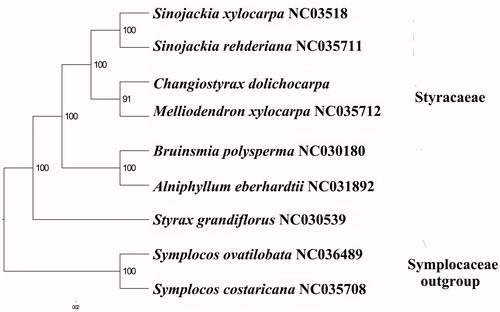
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of six published complete plastome of Styracaceae, using Symplocos ovatilobata and Symplocos costaricana (symplocaceae) outgroups. The phylogenetic analysis indicated that C. dolichocarpa is closer to Melliodendron xylocarpurn than Sinojackia spp. (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of C. dolichocarpa will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Changiostyrax.
Figure 1. The best ML phylogeny recovered from the combined sequences of the 84 coding genes by RAxML. Accession numbers: Sinojackia xylocarpa NC_035418.1, Sinojackia rehderiana NC_035711.1, Changiostyrax dolichocarpa (this study), MH665364, Melliodendron xylocarpum NC_035712.1, Bruinsmia polysperma NC_030180.1, Alniphyllum eberhardtii NC_031892.1, Styrax grandiflorus NC_030539.1, Symplocos ovatilobata NC_036489.1, Symplocos costaricana NC_035708.1.
Acknowledgements
The authors thank Dr. Xiuqun Liu and his students from Huazhong Agricultural University for their help of sampling in the field.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen T. 1995. A new genus of the family from China-Changistyrax. Guangxi Plant. 15:289–292.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Li EH, Wu JQ, Wang Y, Zhao ZE. 2002. A study on the flora of the seed plants from Zigui County of the Three-Gorges Reservoir area. J Wuhan Bot Res. 20:371–380 (in Chinese with English abstract).
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome-DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Qin HN, Yang Y, Dong SY, Qiang H, Yu J, Lina Z, Shengxiang Y, Huiyuan L, Bo L, Yuehong Y, et al. 2017. A list of threatened species of higher plants in China. Biodiversity. 25:697–744.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yao X, Ye Q, Fritsch PW, Cruz BC, Huang H. 2008. Phylogeny of Sinojackia (Styracaceae) based on DNA sequence and microsatellite data: implications for taxonomy and conservation. Ann Bot. 101:651–659.
- Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215: 1235–1248.
