Abstract
The straw-rotting edible fungus Volvariella volvacea is a widely cultivated edible fungus across China and Southeast Asian countries. Three complete mitochondrial genomes of V. volvacea from China, Thailand, and India were determined using the next-generation sequencing technology. The genome sizes of the three strains (China, Thailand, and India) were 62,541 bp, 64,531 bp, and 65,668 bp with GC contents of 38.46%, 38.56%, and 38.52%, respectively. All the genomes encoded 14 conserved protein-coding genes, the small ribosomal RNA subunits (rns), large ribosomal RNA subunits (rnl), and 23 tRNAs were located on the same strand. In the putative protein-coding genes, four introns were distributed in cox1 in the genomes of V23-1 and V8. 5 introns (four introns invaded into cox1and one intron invaded into cob) were detected in Tai8. The phylogenetic analysis confirmed that V. volvacea was a number of Agaricales. This mitochondrial genome may open new avenues for understanding the phylogeny and evolution of Pluteaceae and Agaricales.
Volvariella volvacea (Bull, ex, Fr.) Singer., the Chinese straw mushroom, is an important edible fungus cultivated extensively across China and Southeast Asian countries (Chang and Li Citation1991). According to traditional Chinese medicine, consuming the mushroom is good for the liver and stomach, relieves summer heats, and enriches milk production in women following childbirth (Kalava and Menon 2012; Wang et al. Citation2014). The genome, transcriptome, and some function genes of V. volvacea have been analysed in several studies (Bao et al. Citation2013; Chen et al. Citation2013, Citation2016; Gong et al. Citation2015). However, little is known about the mitochondrial genome of this fungus. In this study, we report three complete mitogenomes of V. volvacea and may provide a phylogenetic analysis of related taxa based on concatenated mitochondrial protein-coding genes.
Three strains provided by Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences were from China, Thailand, and India and named V. volvacea V23-1, Tai8, and V8, respectively. All the monokaryon strains were isolated from dikaryon using the protoplast method (Bao et al. Citation2013) and cultivated under 25 °C on PDA medium for five days. Total DNA extraction, library construction, Illumina sequencing and sequences processing were performed according to the methods previous published (Yang et al. Citation2016; Xu et al. Citation2018). The filtered high quality sequences were assembled into different contigs using A5-miseq 2.0 (Sydney, Australia, Coil et al. Citation2014). And all the assembled contigs were mapped to the database of fungal mitogenomes to extract contigs belonged to the mitogenome of V. volvacea using Blast. The completed mitogenome was annotated using MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl). And neighbour-joining phylogenetic analysis of 26 other species belonged to Agaricomycotina was conducted using MEGA 7.0 (Tokyo, Japan, Kumar et al. Citation2016) ().
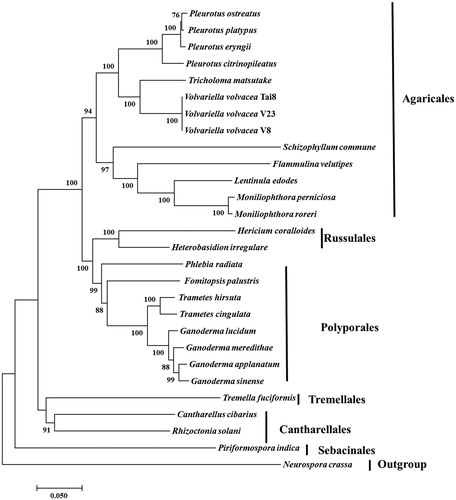
Figure 1. Neighbour-Joining analysis of 28 species belonging to Agaricomycotina (including three V. volvacea strains) based on 13 concatenated amino acid sequences. All the other 25 species used for phylogeny were listed following: Cantharellus cibarius (NC_020368), Flammulina velutipes (NC_021373), Fomitopsis palustris (NC_034349), Hericium coralloides (NC_033903), Ganoderma applanatum (NC_027188), Ganoderma lucidum (NC_021750), Ganoderma meredithae (NC_026782), Ganoderma sinense (NC_022933), Heterobasidion irregulare (NC_024555), Lentinula edodes (NC_018365), Moniliophthora perniciosa (NC_005927), Moniliophthora roreri (NC_015400), Pleurotus citrinopileatus (NC_036998), Pleurotus ostreatus (NC_009905), Pleurotus platypus (NC_036999), Phlebia radiata (NC_020148), Rhizoctonia solani (HF546977), Schizophyllum commune (NC_003049), Serendipita indica (FQ859090), Trametes hirsuta (NC_037239), Tremella fuciformis (NC_036422), Trichosporon asahii var. asahii (MT: JH925097), Trametes cingulata (NC_013933), and Tricholoma matsutake (NC_028135). Neurospora crassa (NC_026614) was served as outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches.
A total of 6232M bp, 1197M bp, and 7790M bp clean data were generated from the sequencing platform for the strains V23-1, Tai8, and V8, respectively. The three circus mitogenomes of V23-1, Tai8, and V8 were 62,541 bp, 64,531 bp, and 65,668 bp in length with GC contents of 38.46%, 38.56%, and 38.52%, respectively. Gene predictions showed 54, 55, and 56 genes determined in V23-1, Tai8, and V8, respectively. Among these genes, 14 conserved protein-coding genes, one small ribosomal RNA subunits (rns) and one large ribosomal RNA subunits (rnl) were detected in all the genomes. The 14 conserved protein-coding genes encoded the three ATP synthases (atp6, apt 8, and apt 9), one apocytochrome b (cob), three cytochrome oxidases (cox1–3), seven subunits of NAD dehydrogenase (nad1-6 and nad4L). Fifteen, 16, and 16 hypothical genes were predicted in V23-1, Tai8, and V8, respectively. The set of 23 tRNA genes could code for all 20 standard amino acids in all the genomes. The four introns were distributed in cox1 but no intron was found in other 13 genes in strains V23-1 and V8. In Tai8, besides four introns invaded into cox1, one intron was detected in cob. Some studies revealed that introns were the main contributors to mitochondrial genome size variations among different strains (Zhang et al. Citation2015).
A total of 13 amino acid sequences were for phylogenetic analysis, including atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad3, nad4, nad4L, nad5, and nad6. The concatenated sequences were aligned using Clustal (Thompson et al. Citation2010). Phylogenetic relationship based on concatenated protein sequences confirmed that V. volvacea was a number of Agaricales and all the three genomes clustered together. Volvariella volvacea clustered together with Tricholoma matsutake belonged to Tricholomataceae. The evolutionarily relationship among Agaricales, Russulales, Polyporales, Cantharellales, and Sebacinales was in accordance with results of previously study (Matheny et al. Citation2006; Garcia-Sandoval et al. Citation2011; Zhao et al. Citation2017). The mitogenomes of V. volvacea would provide new insights into understanding the phylogeny and evolution of Pluteaceae and Agaricales.
Data availability
These genome sequences have been deposited at NCBI (http://www.ncbi.nlm.nih.gov/) under the accession numbers MH647060–MH647062.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bao D, Gong M, Zheng H, Chen M, Zhang L, Wang H, Jiang J, Wu L, Zhu Y, Zhu G, et al. 2013. Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS One. 8:e58294.
- Chang ST, Li SX. 1991. Genetical studies on the sexuality pattern of Volvariella volvacea. In: Van Griensven LJLD, editor. Science and cultivation of edible fungi, USA. Brookfield, VT: Balkema; p. 119–122.
- Chen B, Gui F, Xie B, Deng Y, Sun X, Lin M, Tao Y, Li S. 2013. Composition and expression of genes encoding carbohydrate-active enzymes in the straw-degrading mushroom Volvariella volvacea. PLoS One. 8:e58780.
- Chen B, van Peer AF, Yan J, Li X, Xie B, Miao J, Huang Q, Zhang L, Wang W, Fu J, et al. 2016. Fruiting body formation in Volvariella volvacea can occur independent from it’s a controlled bipolar mating system, enabling homothallic and heterothallic life cycles. G3: Genes Genomes Genetics. 6(7):2135–2146.
- Coil D, Jospin G, Darling AE. 2014. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Garcia-Sandoval R, Wang Z, Binder M, Hibbett DS. 2011. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia. 103:510–524.
- Gong M, Chen MJ, Wang H, Zhu QM, Tan A. 2015. A specific type of cyclin-like F-box domain gene is involved in the cryogenic autolysis of Volvariella volvacea. Mycologia. 107:313–318.
- Kalava SV, Menon SG. 2012. Protective efficacy of the extract of Volvariella volvacea (bulliard ex fries) singer. against carbon tetrachloride induced hepatic injury. Int J Pharm Sci Res. 3:2849–2856.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol.33(7):1870–1874.
- Matheny PB, Curtis JM, Hofstetter V, Aime MC, Moncalvo JM, Ge ZW, Slot JC, Ammirati JF, Baroni TJ, Bougher NL, et al. 2006. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia. 98:982–995.
- Thompson BJD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 2010. The CLUSTAL_X windows interface: exible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 25:4876–4882.
- Wang Y, Chen MJ, Wang H, Wang JF, Bao DP. 2014. Microsatellites in the genome of the edible mushroom, Volvariella volvacea. BioMed Res Int. 2014:1.
- Xu LM, Hinsinger DD, Jiang GF. 2018. The complete mitochondrial genome of the Basidiomycete fungus Pleurotus cornucopiae (Paulet) Rolland. Mitochondrial DNA B. 3:73–75.
- Yang RH, Li Y, Li CH, Xu JP, Bao DP. 2016. The complete mitochondrial genome of the Basidiomycete edible fungus Pleurotus eryngii. Mitochondrial DNA B. 1:772–774.
- Zhang Y, Zhang S, Zhang G, Liu X, Wang C, Xu J. 2015. Comparison of mitochondrial genomes provides insights into intron dynamics and evolution in the caterpillar fungus Cordyceps militaris. Fungal Genet Biol. 77:95–107.
- Zhao RL, Li GJ, Sánchez-Ramírez S, Stata M, Yang ZL, Wu G, Dai YC, He SH, Cui BK, Zhou JL, et al. 2017. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 84:43.
