Abstract
The complete mitochondrial genome was sequenced in two individuals of the surf smelt Hypomesus japonicus. The genome sequences are 16,762 and 16,771 bp in size, and the gene arrangement, composition, and size are very similar to the other smelt mitochondrial genomes published previously. The difference between two H. japonicus genomes studied is 0.37%, which is noticeably higher in comparison with other osmerid fishes. The level of sequence divergence between H. japonicus and related osmerids belonging to genera Hypomeus, Osmerus, and Mallotus varies within a very narrow range (12.31–13.72%) indicating poor phylogenetic resolution of this complex fish group.
The surf smelt Hypomesus japonicus (Brevoort) is a coastal osmerid fish inhabiting the northwestern Pacific Ocean including the Bering Sea, the Sea of Okhotsk, and the Sea of Japan (Pietsch et al. Citation2001). The taxonomy of the genus Hypomesus is highly controversial (Shedko Citation2001 and references therein). Despite intensive investigations based on morphological characteristics (e.g. Berg Citation1949; Klyukanov Citation1970; review in Shedko Citation2001), the species composition and taxonomy of the genus Hypomesus are not clear. The traditional taxonomy based on morphological characteristics is not congruent with the genetic data, which could be explained by the homoplasious morphological characters used in previous studies (Ilves and Taylor Citation2009).
To increase the power of the molecular taxonomy analysis of this complex fish group, we have sequenced two complete mitochondrial (mt) genomes of H. japonicus (GenBank accession numbers MH636616 and MH636617) from the Amur Bay of the Sea of Japan (43°13′17,256″ N; 131°55′37,113″ E; 07.01.2014). The primers were designed with the program mitoPrimer_V1 (Yang et al. Citation2011). The fish specimens are stored at the museum of the National Scientific Center of Marine Biology, Vladivostok, Russia (www.museumimb.ru) under accession numbers MIMB35008 and MIMB35009.
The H. japonicus mt genome sequences are 16,762 and 16,771 bp in size and the gene arrangement, composition, and size are very similar to the smelt fish genomes published previously. We detected 62 single nucleotide and four length differences between the haplotypes 390Hj1 and 392Hj3; total sequence divergence (D xy) was 0.0037 ± 0.0005. The difference between the two H. japonicus mt genomes studied is relatively high in comparison with close species, the European smelt O. eperlanus (D xy = 0.0005 ± 0.0001) and Arctic rainbow smelt Osmerus dentex (D xy = 0.0025 ± 0.0004) (Balakirev et al. Citation2018a, Citation2018b).
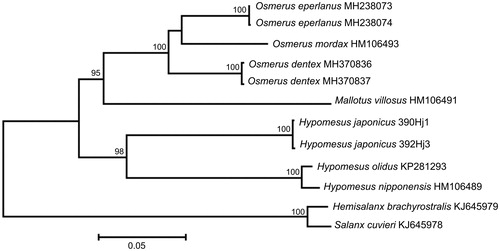
The comparison of the mt genomes now obtained with other complete mt genomes of related groups available in GenBank including genera Hypomesus, Osmerus, Mallotus, Hemisalanx, and Salanx reveals a close affinity of H. japonicus to other Hypomesus species (). The difference between H. japonicus and the cluster of H. nipponensis + H. olidus is high enough (D xy = 0.1269 ± 0.0019) to consider H. japonicus as a separate biological species. However, the difference (D xy) between H. nipponensis and H. olidus is 0.0113 ± 0.0007, which is 8.6 times lower than the average level of divergence (D xy = 0.0971 ± 0.0014) between five available smelts genomes (genera Osmerus, Mallotus, and Hypomesus) excluding H. nipponensis and H. olidus, and which could be explained by erroneous species identification (Balakirev et al. Citation2017) or interspecific replacement of mtDNA (Bernatchez et al. Citation1995). The level of sequence divergence between H. japonicus and other osmerid fishes, H. nipponensis, H. olidus, Osmerus dentex, O. mordax, O. eperlanus, and Mallotus villosus varies in a very narrow range (12.34–13.09%) indicating poor phylogenetic resolution of this group based on the mt sequences; a problem previously discussed (Ilves and Taylor Citation2009; Skurikhina et al. Citation2013).
Figure 1. Maximum likelihood tree for the surf smelt Hypomesus japonicus specimens 390Hj1 and 392Hj3, and GenBank representatives of the order Osmeriformes. The tree is based on the General Time Reversible+ gamma+ invariant sites (GTR + G+I) model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications (values below 75% are omitted).
Disclosure statement
The authors acknowledge no financial interest or benefit from the direct applications of this research. The authors report that they have no conflicts of interest. The research on mitochondrial genome sequencing was conducted at the Department of Ecology and Evolutionary Biology, University of California, Irvine, USA. The data analysis and manuscript preparation were conducted at the National Scientific Center of Marine Biology, Vladivostok, Russia.
Additional information
Funding
References
- Balakirev ES, Saveliev PA, Ayala FJ. 2017. Complete mitochondrial genomes of the Cherskii’s sculpin Cottus czerskii and Siberian taimen Hucho taimen reveal GenBank entry errors: incorrect species identification and recombinant mitochondrial genome. Evol Bioinform. 13:1176934317726783.
- Balakirev ES, Kravchenko AYu, Romanov NS, Ayala FJ. 2018a. Complete mitochondrial genome of the European smelt Osmerus eperlanus (Osmeriformes, Osmeridae). Mitochondrial DNA B: Resour. 3:744–745.
- Balakirev ES, Kravchenko AYu, Romanov NS, Ayala FJ. 2018b. Complete mitochondrial genome of the Arctic rainbow smelt Osmerus dentex (Osmeriformes, Osmeridae). Mitochondrial DNA B: Resour.
- Berg LS. 1949. Freshwater fish of the USSR and adjacent countries. Moscow: Akad. Nauk SSSR.
- Bernatchez L, Glémet H, Wilson CC, Danzmann R. 1995. Introgression and fixation of Arctic charr (Salvelinus alpinus) mitochondrial genome in an allopatric population of brook trout (Salvelinus fontinalis). Can J Fish Aquat Sci. 52:179–185.
- Ilves KL, Taylor EB. 2009. Molecular resolution of the systematics of a problematic group of fishes (Teleostei: Osmeridae) and evidence for morphological homoplasy. Mol Phylogenet Evol. 50:163–178.
- Klyukanov VA. 1970. Morphological basis of the classification of smelts of the genus Hypomesus. Zool J. 49:1534–1542.
- Pietsch TW, Amaoka K, Stevenson DE, MacDonald EL, Urbain BK, López JA. 2001. Freshwater fishes of the Kuril islands and adjacent regions. Species Divers. 6:133–164.
- Shedko SV. 2001. On species composition of smelts of Maritime Territory. Voprosy Ichtiologii. 41:261–264.
- Skurikhina LA, Kukhlevsky AD, Kovpak NE. 2013. Relationships of osmerid fishes (Osmeridae) of Russia: divergence of nucleotide sequences of mitochondrial and nuclear genes. Genes Genom. 35:529–539.
- Yang CH, Chang HW, Ho CH, Chou YC, Chuang LY. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729.
