Abstract
The environmental changes mainly caused by water pollution and human activities have dramatically threatened the survival of the small-scaled Wu’s goby W. polylepis. In the present study, the complete mitogenome of Wuhaniligobius polylepis was determined, which is 16,496 bp in length, containing 13 PCGs, two rRNA genes, 22 tRNA genes and a putative control region. The overall base composition is 28.6%, 29.0%, 26.7%, and 15.7% for A, T, C and G, respectively. The 13 PCGs encode 3,802 amino acids in total, twelve of which use the initiation codon ATG except COI, which uses GTG. In order to explore the systematic status of W. polylepis and further phylogenetic study of Gobiiformes, a maximum-likelihood tree was constructed based on the concatenated sequences of 12 PCGs. The result showed that Pseudogobius, Hemigobius, and Mugilogobius clustered into a clade and then formed a sister-group with W. polylepis, suggesting the invalid former name of M. polylepis or E. polylepis.
The small-scaled Wu’s goby Wuhaniligobius polylepis (Wu and Ni Citation1985), formerly called Mugilogobius polylepis or Eugnathogobius polylepis, distributed in eastern and southern China is found in mangroves and coastal habitats (Nelson Citation2006). The environmental changes caused by water pollution and overfishing, have dramatically threatened the survival of W. polylepis (Huang et al. Citation2013). So far, there are limited molecular studies of this Endangered species, which greatly hinders the phylogenetic studies and further conservation strategies. In order to provide useful molecular information for the restoration strategies and phylogenetic studies for this species, we determined and described the complete mitochondrial genome of W. polylepis and explored the phylogenetic relationship among all the Gobiiformes species, aiming for the phylogenetic studies of Gobiiformes and further management for this species.
The specimen was collected from Beihai, China (N21°29′20″, E109°07′12″) and stored in a refrigerator at −80 °C with accession number 20171016BH02. The total length is 16,496 bp (GenBank No. MG744345). The gene arrangement is identical to that of the typical vertebrate mtDNA (Boore Citation1999; Gong et al. Citation2017a; Gong et al. Citation2017b), including 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a putative control region. Among the 37 genes, nine genes were encoded on the light strand, and the remaining 28 genes were encoded on the heavy strand. The overall base composition is 28.6%, 29.0%, 26.7%, and 15.7% for A, T, C, and G, respectively. The 13 protein-coding genes encode 3,802 amino acids in total. The most frequently used amino acid is Leucine (16.23%), while Cysteine acid (0.76%) is the least frequently used one. Twelve of the PCGs use the initiation codon ATG except for COI, which uses GTG. Seven of them (ND1, ND2, COI, ATP8, ATP6, ND4L and ND5) have TAA as the stop codon, while ND4 is inferred to termination with AGG, ND3 and ND6 with TAG, and the remaining four (COII, COIII, and Cyt b) are inferred to termination with an incomplete stop codon T.
The major non-coding sequence or control region (CR) is commonly located between the tRNA-Pro and tRNA-Phe genes. It is 839 bp in length and is characterized by a relatively high AT content (65.4%). The base composition is 32.1%, 33.3%, 19.2%, and 15.5% for A, T, C, and G, respectively. Like that of other vertebrates (Guo et al. Citation2003; Ragauskas et al. Citation2014; Wang et al. Citation2014; Gong et al. Citation2015), the CR of this species is also partitioned into three domains and some similar conserved-sequence blocks are found, including the termination associated sequence (TAS), the central conserved sequence blocks (CSB-F, D, C and A) and the conserved sequence blocks (CSB-2 and 3).
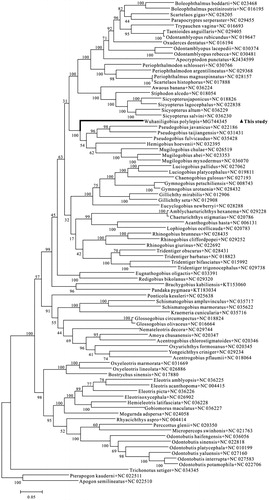
In order to explore the systematic status of W. polylepis and carry out further phylogenetic study of Gobiiformes, a maximum-likelihood tree was constructed based on the concatenated sequences of 12 PCGs. The tree clearly showed that Pseudogobius, Hemigobius, and Mugilogobius are clustered into a clade and form a sister-group with W. polylepis, suggesting that the former name of M. polylepis or E. polylepis () was invalid.
Figure 1. Maximum Likelihood (ML) tree of Gobiiformes species based on 12 PCGs. The bootstrap values are based on 500 resamplings. The number at each node is the bootstrap probability. The number after the species name is the GenBank accession number. The genome sequence in this study is labeled with triangle.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Gong L, Lu Z-M, Guo B-Y, Ye Y-Y, Liu L-Q. 2017a. Characterization of the complete mitochondrial genome of the tidewater goby, Eucyclogobius newberryi (Gobiiformes; Gobiidae; Gobionellinae) and its phylogenetic implications. Conserv Genet Resour. 10:93–97.
- Gong L, Liu L-Q, Guo B-Y, Ye Y-Y, Lü Z-M. 2017b. The complete mitochondrial genome characterization of Thunnus obesus (Scombriformes: Scombridae) and phylogenetic analyses of Thunnus. Conserv Genet Resour. 9:379–383.
- Gong L, Shi W, Si LZ, Wang ZM, Kong XY. 2015. The complete mitochondrial genome of peacock sole Pardachirus pavoninus (Pleuronectiformes: Soleidae) and comparative analysis of the control region among 13 soles. Mol Biol. 49:408–417.
- Guo X, Liu S, Liu Y. 2003. Comparative analysis of the mitochondrial DNA control region in cyprinids with different ploidy level. Aquaculture. 224:25–38.
- Huang S-P, Zeehan J, Chen I-S. 2013. A new genus of Hemigobius generic group goby based on morphological and molecular evidence, with description of a new species. J Marine Sci Technol. 21:146–155.
- Nelson JS. 2006. Fishes of the word. 4th ed. New York: John Wiley.
- Ragauskas A, Butkauskas D, Sruoga A, Kesminas V, Rashal I, Tzeng W-N. 2014. Analysis of the genetic structure of the European eel Anguilla anguilla using the mtDNA D-loop region molecular marker. Fish Sci. 80:463–474.
- Wang S-Y, Shi W, Miao X-G, Kong X-Y. 2014. Complete mitochondrial genome sequences of three rhombosoleid fishes and comparative analyses with other flatfishes (Pleuronectiformes). Zool Stud. 53:80.
- Wu HL, Ni Y. 1985. On two new species of Mugilogobius Smit (Perciformes: Gobiidae) from China. Zool Res. 6:93–98 [In Chinese].
