Abstract
In this study, the complete mitochondrial genome (mitogenome) of Exopalaemon annandalei was amplified and analyzed. The mitogenome is 15,718 bp in length, encoding 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region (CR). The nucleotide frequency of the mitogenome is as follows: A, 34.81%; C, 23.24%; G, 12.68%; and T, 29.25%. Seven kinds of the initiation codon and five kinds of termination codon are employed in the 13 PCGs. Phylogenetic analysis show E. annandalei to be in sister-relationship with E. carinicauda. The complete mitogenome sequence information of E. annandalei would play an important part in further studies on molecular systematics and phylogeny.
Exopalaemon annandalei, a common small-scale economic shrimp, is a unique species that is mainly distributed along coastal areas in China (Zhuang et al. Citation2008), with the characteristic of rapid propagation (Wu Citation1993). In view of the importance of E. annandalei, we sequenced its mitochondrial genome and used this to clarify its phylogenetic position.
In this study, the samples of E. annandalei (accession number: B0012-1) were collected from the East China Sea (121°59′10″E, 31°04′48″N). The specimens now were stored in East China Sea Environmental Monitoring Center. Partial sequences of COI and 16S rRNA genes were amplified by PCR using universal primers. Based on these two PCR products, we designed two specific primer pairs to amplify two DNA fragments of the mitogenome of E. annandalei, respectively. Subsequently, these two PCR products were sequenced using Shot Gun Sequencing. Then all the sequences obtained were analyzed and assembled.
The complete mitogenome of E. annandalei (GenBank number MG787410) is a closed circular molecule, 15,718 bp in length. The gene content is similar to other animal mitochondrial genomes (Zhang et al. Citation2017), including 22 tRNA genes, 13 PCGs (COX1-3, NAD1-6, and 4L, COb, ATP6 and ATP8), two mitochondrial ribosomal RNAs (12S and 16S rRNA), and a control region (CR). Nucleotide frequency of the mitogenome is as follows: A, 34.81%; C, 23.24%; G, 12.68%; and T, 29.25%. The base composition has A + T bias, with the value of 67.07%. The ND1, ND4, ND4L, ND5, 16S rRNA, 12S rRNA, tRNA-Cys, tRNA-His, tRNA-Gln, tRNA -Leu, tRNA-Phe, tRNA-Pro, tRNA-Tyr, and tRNA-Val genes are encoded on the reverse strand of the mitogenome, whereas the remaining genes are encoded on the forward strand. Many different kinds of initiation codons exist in the 13 PCGs: six PCGs (ATP6, COX2, COX3, ND1, ND4L, and Cob) start with the ATG typical initiation codon, ND2 and ND5 genes share the ATA initiation codon, and the remaining five genes use ACG, ATC, ATT, GTG, and TTG as initiation codons. Both complete and incomplete stop codons are used in the protein-coding genes: COX1, COX2, and COb genes have incomplete stop codons T–/TA-, while the remaining genes have the stop codon TAA/TAG/TAC. All 22 tRNAs were identified in the E. annandalei mitogenome and range from 61 bp to 70 bp in length. The CR (934 bp) of the E. annandalei mitogenome is located between 12S rRNA and tRNA-Ile. We found microsatellite-like repeat (AT) 5 and (TA) 5 elements in the CR.
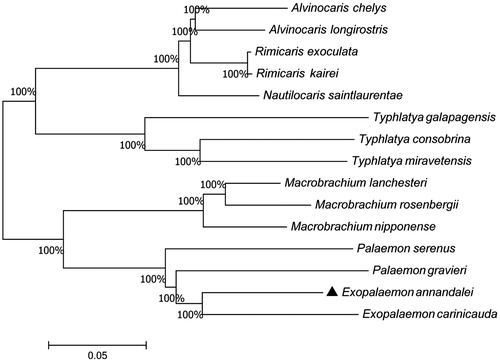
To clarify the phylogenetic relationship among decapoda, a phylogenetic tree was constructed using the Neighbor-Joining method () (Kumar et al. Citation2016). E. annandalei is grouped with E. carinicauda as sister species and located in the basal branch. Palaemon gravieri (Kim et al. Citation2017) and Palaemon serenus (Gan et al. Citation2016) have a close relationship with E. annandalei. This study would provide basic data for research on genetic analyses and evolutionary status of these species in the future.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Clayton DA. 1992. Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 141:217–232.
- Gan HY, Gan HM, Lee YP, Austin CM. 2016. The complete mitogenome of the rock pool prawn Palaemon serenus (Heller, 1862) (Crustacea: Decapoda: Palaemonidae). Mitochondrial DNA B. 27:3155–3156.
- Kim ST, Kim G, Yoon TH, Kim AR, Kim HJ, Kim HW. 2017. Complete mitochondrial genome of Palaemon gravieri (Yu, 1930) (Crustacea: Decapoda: Palaemonidae). Mitochondrial DNA B. 28:277–278.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Wu CW. 1993. On the biology and ecological distribution of shrimp, Palaemon (exopalaemon) annandalei,at hangzhou bay. J Zhejiang Ocean Univ. 12:21–31.
- Zhang XC, Li W, Zhao J, Chen HG, Zhu XP. 2017. The complete mitogenome of Pelochelys cantorii (Guangning) and the comparative analysis of different habitats. Conserv Genet Resour. 10:1–4.
- Zhuang P, Song C, Zhang LZ. 2008. Comparison of nutritive components of Exopalaemon annandalei and Macrobrachium nipponensis collected from the Yangtze Estuary. Acta Zool Sin. 54:822–829.