Abstract
The first complete chloroplast genome sequence for Narcissus is assembled and annotated in this study. The total length of the N. poeticus chloroplast genome is 160,099 bp and comprises the large single copy (LSC) spanning 86,445 bp, the small single copy (SSC) spanning 16,434 bp, and two inverted repeat regions each of 28,610 bp length. The truncated copy of ycf1 before the junction between IRB and SSC was 1277–2428 bp longer than in other included Asparagales samples. A potential pseudogene, cemA, was also identified. This is the first reported plastome for Amaryllidaceae subfamily Amaryllidoideae.
Keywords:
Narcissus L. is a member of Amaryllidaceae, a family comprising several horticulturally important plant genera (Heywood et al. Citation2007). Narcissus poeticus is the type species of the genus, however, its placement among other daffodils remains unresolved (Marques et al. Citation2017). Currently, there are no published daffodil chloroplast genomes and only a few genomes are available for Amaryllidaceae, none for subfamily Amaryllidoideae. Here we report the complete chloroplast genome sequence of N. poeticus to address this and to identify new regions of genomic variability.
Leaf material was collected in silica gel from N. poeticus grown at RHS Garden Wisley, UK (51.312695° N, 0.476724° W). Herbarium voucher specimen was deposited at WSY (WSY0108940). Total genomic DNA was extracted using the QIAGEN DNeasy Plant Mini Kit (QIAGEN, Manchester, UK). Library development and 150 bp PE sequencing on 1/16 of an Illumina HiSeq 4000 lane were done at the Oxford Genomics Centre (Oxford, UK). The chloroplast genome was assembled with Fast-Plast v1.2.6 (McKain and Wilson Citation2017) and NovoPlasty v2.7.0 (Dierckxsens et al. Citation2017). Fast-Plast assemblies were run with all 38 M reads. Reads were trimmed to remove NEB-PE adapter sequences. Bowtie reference index was built with Asparagales chloroplast genomes included in Fast-Plast. For the NovoPlasty assembly, adapters were trimmed with Trimmomatic v0.36 (Bolger et al. Citation2014) using the same adapter sequences. A ndhF sequence of N. poeticus (KT124416) was used as the starting seed and memory was limited to 8 Gb. All other parameters were unchanged. Junctions of the inverted repeats were confirmed by Sanger sequencing.
The complete chloroplast genome was annotated using Verdant (McKain et al. Citation2017) and corrected by comparing it with published annotations (Hesperoyucca whipplei – KX931459; Hosta ventricosa – KX931460; Yucca schridigera – KX931469; Cocos nucifera – KF285453). The N. poeticus cpDNA genome sequence was aligned with 13 published Asparagales plastome sequences using MAFFT (Katoh and Standley Citation2013). A maximum likelihood estimate was conducted with RAxML v8.2.11 (Stamatakis Citation2014) within Geneious v11.1.5 (http://www.geneious.com, Kearse et al. Citation2012) using model GTR + G and 1000 bootstrap replicates.
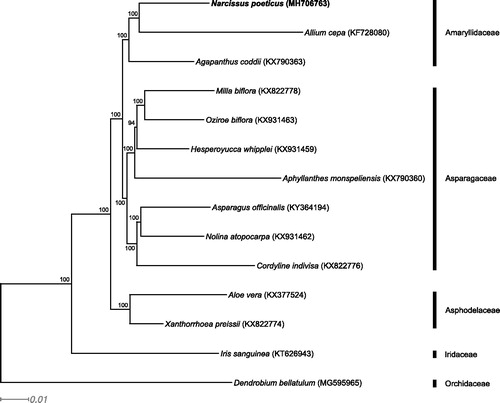
The chloroplast genome sequence of N. poeticus (MH706763) is 160,099 bp, comprises the large single copy (LSC) spanning 86,445 bp, the small single copy (SSC) spanning 16,434 bp, and two inverted repeat regions each of 28,610 bp length. The junction between IRB and SSC (JSB) is 34 bp within the ndhF gene. The junction between SSC and IRA (JSA) is within the ycf1 gene, which is 5,346 bp long, of which 2737 bp lies in the inverted repeat. Therefore, there is a 2737 bp long truncated version of ycf1 at JSB. This is 1277–2428 bp longer than the truncated copy of ycf1 in other Asparagales sequences analyzed here, identified by IRScope (Amiryousefi et al. Citation2018). A poly-A region, at the start of cemA shifts the ORF potentially inactivating this gene. The Amaryllidaceae samples form a clade with N. poeticus sister to Allium cepa () topologically consistent with Seberg et al. (Citation2012).
Figure 1. RAxML output tree with bootstrap consensus values based on 14 complete chloroplast genome sequences. The numbers at each node indicate bootstrap support. GenBank accession numbers are given in brackets. Text in bold shows the chloroplast genome developed in this study. Families of the sampled taxa are shown on the right.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34:3030–3031.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Heywood VH, Brummit RK, Culham A, Seberg O. 2007. Amaryllidaceae. Flowering plant families of the world. Kew: Royal Botanic Gardens.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Marques I, Aguilar JF, Martins-Louçao MA, Moharrek F, Feliner GN. 2017. A three-genome five-gene comprehensive phylogeny of the bulbous genus Narcissus (Amaryllidaceae) challenges current classifications and reveals multiple hybridization events. Taxon. 66:832–854.
- McKain MR, Hartsock RH, Wohl MM, Kellogg EA. 2017. Verdant: automated annotation, alignment and phylogenetic analysis of whole chloroplast genomes. Bioinformatics. 33:130–132.
- McKain MR, Wilson M. 2017. Fast-Plast: rapid de novo assembly and finishing for whole chloroplast genomes. Version 1.2.6.
- Seberg O, Petersen G, Davis JI, Pires JC, Stevenson DW, Chase MW, Fay MF, Devey DS, Jørgensen T, Sytsma KJ, Pillon Y. 2012. Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. Am J Bot. 99:875–889.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
