Abstract
The complete nucleotide sequence of the mitochondrial genome of the bambusicolous fungus Fusarium bambusae was determined using the next-generation sequencing technology. The circular molecule is 63,593 bp long with a GC content of 31.92%. Gene prediction revealed 44 genes encoding 15 conserved proteins, 27 tRNAs, and the large and small ribosomal RNAs. All genes are located on the same strand. The tRNA genes contain codons for all 20 standard amino acids. It turns out to be similar to the previously sequenced mitochondrial genomes of Fusarium circinatum and F. verticillioides. The differences lie in the number of introns embodied in protein-coding genes. Four introns exist in the mitochondrial genome of F. verticillioides, 10 in F. bambusae, and 14 in F. circinatum. The phylogenetic analysis confirmed F. bambusae as a member of Fusarium (Nectriaceae). The mitochondrial genome of F. bambusae will contribute to the understanding of phylogeny and evolution of the genus and family.
Fusarium bambusae is a bambusicolous fungus discovered on culm of bamboo or rotten bamboo in eastern China, including Anhui, Henan, and Zhejiang provinces. It is characterized by soft-textured dark purple ascomata, two-layered perithecial wall, and clavate asci with 4–8 ascospores (Zhang and Zhuang Citation2003; Zhuang Citation2013). Formerly, this fungus was known as Gibberella bambusae, but recently transferred to Fusarium (Zeng and Zhuang Citation2017) according to genera in Bionectriaceae, Hypocreaceae, and Nectriaceae proposed for acceptance or rejection (Rossman et al. Citation2013). Its life circle and applicability have not been well investigated.
The F. bambusae strain 5137 was isolated from an ascoma living on rotten bamboo from the Jigong Mountain (N31°49′, E114°04′), Henan Province, China. The specimen is deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS 86476). DNA extraction, library construction, and sequencing were performed as described in the previous studies (Wang et al. Citation2016, Citation2017). The 125 bp pair-end reads were assembled using CLC Genomics Workbench (Version 8.0.3; CLC Bio, Aarhus, Denmark). The mitochondrial genome of Trichoderma reesei (NC_003388) was served as a reference to identify the assembled scaffolds belonging to mitochondrial genome. After filtering with BLAST, only one scaffold was obtained. Manual comparison of the ends of the scaffold helped to link them into a circular molecule. This mitochondrial genome was annotated using MFannot (Lang et al. Citation2014). Ten mitochondrial genomes belonging to Nectriaceae and Bionectriaceae released on the public database were included in the Neighbour-joining phylogenetic analysis using MEGA6 (Tamura et al. Citation2013).
The complete sequence of F. bambusae mitochondrial genome (GenBank accession number MH684411) is 63,593 bp long with the GC content of 31.92%. It encodes 15 conserved proteins, 27 tRNAs, and the large and small ribosomal RNAs. All structural genes are located on the same strand. The tRNA genes contain codons for all 20 standard amino acids. Most amino acids are represented by only one tRNA gene; however, two trnL (trnL-UAA and trnL-UAG), two trnS (trnS-GCU and trnS-UGA), three trnM-CAU and four trnR (trnR-ACG, trnR-UCG, and double copies of trnR-UCU) genes are found in this mitochondrial genome. Eleven introns are detected in five genes, i.e. each in rnl, nad2, and cox2, three in cob and five in cox1.
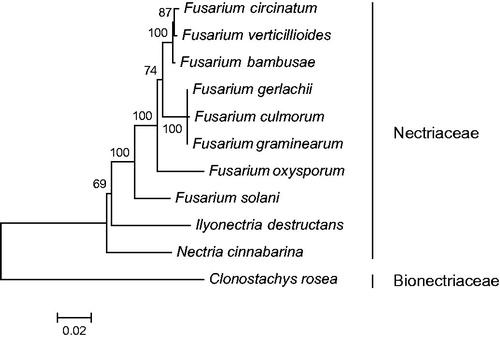
As shown in , 10 members of Nectriaceae are included in the phylogenetic analysis, including eight taxa of Fusarium. Fusarium circinatum (NC_022681) and F. verticillioides (NC_016687) are determined as sisters of F. bambusae with strong support. The three share the same composition and order of protein, rRNA and tRNA genes. However, they differ in the number of introns embodied in protein-coding genes. Four introns exist in the mitochondrial genome of F. verticillioides, 10 in F. bambusae, whereas 14 in F. circinatum. The mitochondrial genome of F. bambusae will contribute to the understanding of phylogeny and evolution of Nectriaceae.
Figure 1. Phylogenetic relationship of 10 taxa of Nectriaceae (Hypocreales, Ascomycota) determined by Neighbour-joining analysis based on concatenated sequences of 15 translated mitochondrial proteins. The 15 proteins included subunits of the respiratory chain complexes (cob, cox1, cox2, cox3), ATPase subunits (atp6, atp8, and atp9), NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, nad6), and ribosomal protein S3 (rps3). The concatenated sequences were aligned using MAFFT. The following mitogenomes were used in this analysis: Fusarium circinatum (NC_022681), F. culmorum (NC_026993), F. gerlachii (NC_025928), F. graminearum (NC_009493), F. oxysporum (NC_017930), F. solani (NC_016680), F. verticillioides (NC_016687), Ilyonectria destructans, and Nectria cinnabarina (NC_030252). Clonostachys rosea (NC_036667, Bionectriaceae) was served as outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Bootstrap values no less than 50% are shown.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Lang BF, Jakubkova M, Hegedusova E, Daoud R, Forget L, Brejova B, Vinar T, Kosa P, Fricova D, Nebohacova M, et al. 2014. Massive programmed translational jumping in mitochondria. Proc Natl Acad Sci USA. 111:5926–5931.
- Rossman AY, Seifert KA, Samuels GJ, Minnis AM, Schroers H-J, Lombard L, Crous PW, Põldmaa K, Cannon PF, Summerbell RC, et al. 2013. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus. 4:41–51.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang XC, Zeng ZQ, Zhuang WY. 2016. The complete mitochondrial genome of the important phytopathogen Nectria cinnabarina (Hypocreales, Ascomycota). Mitochondrial DNA Part A. 27:4670–4671.
- Wang XC, Zeng ZQ, Zhuang WY. 2017. The complete mitochondrial genome of the important mycoparasite Clonostachys rosea (Hypocreales, Ascomycota). Mitochondrial DNA Part B. 2:180–181.
- Zeng ZQ, Zhuang WY. 2017. Eight new combinations of Bionectriaceae and Nectriaceae. Mycosystema. 36:278–281.
- Zhang XM, Zhuang WY. 2003. Re-examinations of Bionectriaceae and Nectriaceae (Hypocreales) from temperate China on deposit in HMAS. Nova Hedwigia. 76:191–200.
- Zhuang WY, editor. 2013. Flora Fungorum Sinicorum vol. 47 Nectriaceae et Bionectriaceae. Beijing, China: Science Press. Chinese.
