Abstract
The complete mitochondrial genome of Metopograpsus quadridentatus was determined to be 15,520 bp in length. It consists of 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs and a control region. There are 13 overlapping regions in the genome with 1 to 25 bp length. The largest overlapping region is located between nad1 and trnL1. The AT-skew and GC-skew for the whole mitogenome are both negative, indicating a higher occurrence of Ts than As and Cs than Gs. The molecular data here we presented could play a useful role to study the evolutionary relationships and population genetics of Grapsidae crabs.
Metopograpsus quadridentatus, belongs to the genus Metopograpsus, mainly lives in mangroves and intertidal areas of the eastern Indian and West Pacific oceans, from Singapore to southern China (Ng et al. Citation2008). The genus Metopograpsus (H. Milne Edwards, 1853) belongs to the family Grapsidae (within Crustacea: Decapoda: Thoracotremata : Brachyura) and includes six species of intertidal crabs from sheltered rocky shores or mangroves and associated muddy areas (Paulay Citation2007). The taxonomy of the genus Metopograpsus has been questioned due to minor diagnostic morphological differences among species (Fratini et al. Citation2018). Therefore, to facilitate the future researches of taxonomic resolution, population genetic structure and phylogeography, the complete mitochondrial genome of M. quadridentatus was determined.
The samples were collected from the coastal intertidal zone of Shanghai (China) on 19 September 2017 (121°54'38″E, 30°51'28″N). All samples were stored at the Jiangsu Provincial Key Laboratory of Coastal Wetland Bioresources and Environmental Protection, Yancheng Teachers University, Yancheng, Jiangsu Province, China. Total DNA was extracted from the muscle tissue and using the Aidlab Genomic DNA Extraction Kit (Aidlab Biotech, Beijing, China). The mitogenomes of M. quadridentatus were sequenced by next-generation sequencing (Illumina HisSeq 4000), and clean data without sequencing adapters were de novo assembled by the NOVOPlasty software (Dierckxsens et al. Citation2017). The mitogenome of M. quadridentatus is a closed circular molecule 15,520 bp in size. The gene content is typical of Decapoda mitochondrial genomes, including 13 PCGs (cox1–3, nad1–6, nad4L, cob, atp6 and atp8), 2 rRNA genes (rrnS and rrnL), 22 tRNA genes and a major non-coding region known as the CR. Twenty-three genes are encoded on the heavy (+) strand while the remaining 14 genes (4 of the 13 PCGs, 8 tRNAs and 2 rRNAs) are located on the light (−) strand. There are 13 overlapping regions in the genome with 1 to 25 bp length. The genome shows 19 intergenic sequences varying from 1 to 80 bp in size. The AT-skew and GC-skew for the whole mitogenome are both negative, indicating a higher occurrence of Ts than As and Cs than Gs. The mitogenome of M. quadridentatus has been deposited in GenBank under accession number MH310445.
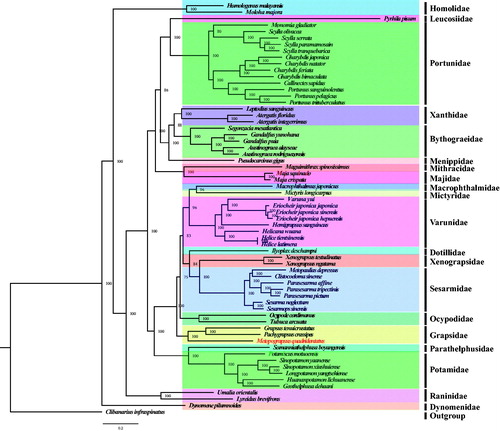
To reconstruct the phylogenetic relationship among crabs, the complete mitogenomes of other 61 Brachyura and one outgroup (Clibanarius infraspinatus) were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). The concatenated set of nucleotide sequences were used for phylogenetic analysis, which was performed with the BI and ML methods using MrBayes v3.2.6 (Huelsenbeck and Ronquist Citation2001) and RaxML (Stamatakis Citation2014), respectively. In this study, both the BI and ML analyses showed that each family in the tree formed a monophyletic clade (). From the phylogenetic tree, we found M. quadridentatus and (Grapsus tenuicrustatus + Pachygrapsus crassipes) are clustered in one branch with high nodal support value (BI posterior probabilities [PP] = 100, ). Additionally, the relationship among Sesarmidae, Xenograpsidae, Grapsidae, Varunidae, Potamidae, Bythograeidae, and Portunidae is also identified and is largely consistent with previous researches (Shen et al. Citation2013; Wang et al. Citation2018a,Citationb).
Figure 1. Phylogeny of Brachyura based on nucleotide sequences. The phylogenetic tree was inferred from the nucleotide sequences of 13 mitogenome PCGs using BI and ML methods. Numbers on branches indicate posterior probability (BI). Clibanarius infraspinatus was used as outgroup. The genbank accession numbers for all of the sequences is listed as follows: Atergatis floridus NC_037201.1, Atergatis integerrimus NC_037172.1, Austinograea alayseae NC_020314.1, Austinograea rodriguezensis NC_020312.1, Callinectes sapidus NC_006281.1, Charybdis feriata NC_024632.1, Charybdis japonica NC_013246.1, Charybdis natator NC_036132.1, Clibanarius infraspinatus NC_025776.1, Clistocoeloma sinense NC_033866.1, Dynomene pilumnoides KT182070.1, Eriocheir japonica hepuensis NC_011598.1, Eriocheir japonica japonica NC_011597.1, Eriocheir japonica sinensis NC_006992.1, Gandalfus puia NC_027414.1, Gandalfus yunohana NC_013713.1, Geothelphusa dehaani NC_007379.1, Grapsus tenuicrustatus NC_029724.1, Charybdis bimaculata MG489891.1, Helicana wuana NC_034995.1, Helice latimera NC_033865.1, Helice tientsinensis NC_030197.1, Hemigrapsus sanguineus NC_035307.1, Homologenus malayensis NC_026080.1, Huananpotamon lichuanense NC_031406.1, Ilyoplax deschampsi NC_020040.1, Leptodius sanguineus NC_029726.1, Longpotamon yangtsekiense NC_036946.1, Lyreidus brevifrons NC_026721.1, Macrophthalmus japonicus NC_030048.1, Maguimithrax spinosissimus NC_025518.1, Maja crispata NC_035424.1, Maja squi-nado NC_035425.1, Metopaulias depressus NC_030535.1, Mictyris longicarpus NC_025325.1, Moloha majora NC_029361.1, Monomia gladiator NC_037173.1, Ocypode cordimanus NC_029725.1, Potamiscus motuoensis KY285013.1, Pachygrapsus crassipes NC_021754.1, Parasesarma affine MH310444, Parasesarma pictum MG580780, Parasesarma tripectinis NC_030046.2, Portunus pelagicus NC_026209.1, Portunus sanguinolentusNC_028225.1, Portunus trituberculatus NC_005037.1, Pseudocarcinus gigas NC_006891.1, Pyrhila pisum NC_030047.1, Scylla olivacea NC_012569.1, Scylla paramamosain NC_012572.1, Scylla serrata NC_012565.1, Scylla tranquebarica NC_012567.1, Segonzacia mesatlanticaNC_035300.1, Sesarma neglectum NC_031851.1, Sesarmops sinensis NC_030196.1, Sinopotamon xiushuiense NC_029226.1, Sinopotamon yaanense NC_036947.1, Somanniathelphusa boyangensis NC_032044.1, Tubuca arcuata KX911977.1, Umalia orientalis NC_026688.1, Varuna yui NC_037155.1, Xenograpsus ngatama NC_035951.1, Xenograpsus testudinatus NC_013480.1.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. The authors report that they have no competing interest.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Fratini S, Cannicci S, Schubart CD. 2018. Molecular phylogeny of the crab genus Metopograpsus H. Milne Edwards, 1853 (Decapoda : Brachyura : Grapsidae) reveals high intraspecific genetic variation and distinct evolutionarily significant units. Invert Systematics. 32:215–223.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Ng PKL, Guinot DG, Davie PJF. 2008. Systema Brachyurorum: part I. An annotated checklist of extant Brachyuran crabs of the world. Raffles B Zool. 17:1–286.
- Paulay G. 2007. Metopograpsus oceanicus (Crustacea: Brachyura) in Hawai'i and Guam: another recent invasive? Pac Sci. 61:295–300.
- Shen H, Braband A, Scholtz G. 2013. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mole Phyl Evol. 66:776–789.
- Stamatakis A. 2014. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 30:1312–1313.
- Wang Z, Shi X, Tao Y, Wu Q, Bai Y, Guo H, Tang D. 2018a. The complete mitochondrial genome of Parasesarma pictum (Brachyura : Grapsoidea : Sesarmidae) and comparison with other Brachyuran crabs. Genomics. 1-11. doi: 10.1016/j.ygeno.2018.05.002.
- Wang Z, Wang Z, Shi X, Wu Q, Tao Y, Guo H, Ji C, Bai Y. 2018b. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int J Biol Macromol. 118:31–40.
