Abstract
Here, the complete Gracilaria tenuistipitata var. liui mitogenome was determined and analyzed. The complete G. tenuistipitata var. liui mitogenome length was 25,879 bp and contained 50 genes including 24 protein-coding, 2 rRNA, and 23 tRNA genes and one unidentified open reading frame (ORF). Of the 24 protein-coding genes, 23 (95.83%) terminated with the stop codon TAA, and one (4.17%) with TAG (rps3 gene). All protein-coding genes in G. tenuistipitata var. liui started with ATG codon. Mitogenome phylogenetic analysis revealed that G. tenuistipitata var. liui firstly clustered together with Gracilaria tenuistipitata. The complete mitogenome sequence would help in understanding Gracilaria evolution.
Gracilaria tenuistipitata var. liui Zhang and Xia (Gracilariaceae, Rhodophyta) is found locally in South-East Asia and China (Phang Citation2006) and extensively cultivated in southern China (Chiang Citation1980). Being the sources for the phycocolloids and food, the marine red alga Gracilaria tenuistipitata var. liui Zhang and Xia is economically important (Oliveira et al. Citation2000). To date, many studies have been performed on the growth of G. tenuistipitata var. liui (Israel et al. Citation1999; Li-hong et al. Citation2002; Xu et al. Citation2009). However, information on its genetics and systematics is limited. Herein, we determined the complete mitogenome of G. tenuistipitata var. liui to provide new molecular data for genetics study.
Gracilaria tenuistipitata var. liui individual (specimen number: 2016110001) was collected from Shantou, Guangdong Province in the eastern China (23°24′19′′N, 117°3′15′′E). The high-throughput sequencing method and data processing followed Liu et al. (Citation2017). Gracilaria salicornia (GenBank accession number: NC_023784) was used as the seed sequence.
The complete G. tenuistipitata var. liui (MG592728) mitogenome comprised a circular DNA molecule with the length of 25,879 bp. The overall A + T content of the complete mitogenome was 72.9%. The mitogenome contained 50 genes, including 24 protein-coding, two rRNA, 23 tRNA genes, and one unidentified open reading frame (ORF). Of the 24 protein-coding genes, 23 (95.83%) terminated with the TAA stop codon, and one (4.17%) with TAG (rps3 gene)). All protein-coding genes in G. tenuistipitata var. liui were found to have the start codon ATG. The lengths of two rRNA genes were 2,619 bp (rnl) and 1,394 bp (rns) respectively.
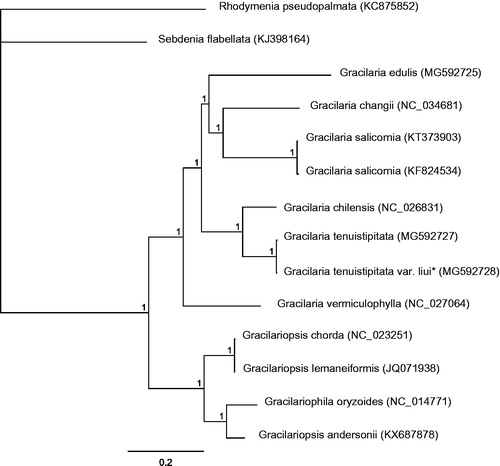
Bayesian analysis based on the complete mitogenomes of the twelve Gracilariaceae species was conducted using MrBayes v. 3. 1.2 (Huelsenbeck and Ronquist Citation2001). Rhodymenia pseudopalmata (KC875852) and Sebdenia flabellata (KJ398164) served as the out-group. All algae were divided into two clades: Gracilaria and Gracilariopsis (). Phylogenetic analyses showed that Gracilaria tenuistipitata var. liui firstly clustered with Gracilaria tenuistipitata. The complete mitogenome data provided in this work would help us to understand Gracilaria evolution.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Chiang YM. 1980. Cultivation of gracilaria (Rhodophycophyta, Gigartinales) in Taiwan. In: Levring T, editor. Xth International Seaweed Symposium. Berlin: Walter de Gruyter & Co.; p 569–574.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England). 17:754–755.
- Israel A, Martinez-Goss M, Friedlander M. 1999. Effect of salinity and pH on growth and agar yield of Gracilaria tenuistipitata var. liui in laboratory and outdoor cultivation. J Appl Phycol. 11:543–549.
- Li-hong H, Madeline W, Pei-yuan Q, Ming-yuan Z. 2002. Effects of co-culture and salinity on the growth and agar yield of Gracilaria tenuistipitata varliui Zhanget Xia. Chin J Oceanol Limnol. 20:365–370.
- Liu N, Wang GL, Li Y, Zhang L, Meinita MDN, Chen WZ, Liu T, Chi S. 2017. The complete mitochondrial genome of the economic red alga, Gracilaria chilensis. Mitochondr DNA B Res. 2:716–717.
- Oliveira EC, Alveal K, Anderson RJ. 2000. Mariculture of the agar-producing gracilarioid red algae. Rev Fish Sci. 8:345–377.
- Phang SM. 2006. Seaweed resources in Malaysia: current status and future prospects. Aqua Ecosys Health Manag. 9:185–202.
- Xu Y, Wei W, Fang J. 2009. Effects of salinity, light and temperature on growth rates of two species of Gracilaria (Rhodophyta). Chin J Oceanol Limnol. 27:350.