Abstract
In this study, we obtained the complete plastid genome of Grateloupia filicina (Lamouroux) C. Agardh using the high-throughput sequencing method. It had circular mapping organization with the length of 195,990 bp and contained 265 genes including 195 protein-coding genes, three rRNA genes, one tmRNA gene, 28 tRNA genes and 38 unidentified open reading frames (ORFs). Moreover, phylogenetic analysis revealed that G. filicina firstly clustered with Grateloupia taiwanensis. The complete plastid genome obtained in this work would be useful for further understanding the evolution of Grateloupia.
Red alga Grateloupia filicina (Lamouroux) C. Agardh (Halymeniales, Rhodophyta) is an edible marine macroalga which is distributed globally from tropical to warm temperate regions (Wynne Citation1998; Masuda et al. Citation2000). In recent years, G. filicina has attracted us in a considerable attention owing to its multiple biological functions. The sulphated polysaccharide extracted from G. filicina has been reported to have high anticoagulant activity (Nikapitiya et al. Citation2007), anti-HIV-1 activity (Wang et al. Citation2007), antiviral activity (Sun et al. Citation2017) and antiangiogenic effects (Yu et al. Citation2012). Here, we determined the complete plastid genome of G. filicina (GenBank accession number: MG598531) and constructed phylogenetic analysis to provide new molecular information.
G. filicina samples (specimen number: 2016030029) were collected from Xiangshan Harbor, Zhejiang Province in eastern China (29°30′20′N, 121°35′6′′E) and deposited in the Culture Collection of Seaweed at the Ocean University of China. Paired-end reads were sequenced using the HiSeq × Ten system (Illumina, USA). Approximately 9 Gb of paired-end (150 bp) clean reads were randomly retrieved from the total sequencing output and used as input into NOVOPlasty (Dierckxsens et al. Citation2017) to assemble the plastid genome. The plastid genome of Grateloupia taiwanensis (GenBank accession number: KM999231) was used as the reference sequence. Transfer RNA genes were identified using the tRNAscan-SE Search Server (Schattner et al. Citation2005). Other regions were annotated by comparison with those of G. taiwanensis by using Geneious R10 (Biomatters Ltd., Auckland, New Zealand). Phylogenetic analysis based on rbcL and 16S genes from 17 Rhodymeniophycidae plastid genomes were conducted using Maximum likelihood (ML) method. ML tree search and ML bootstrap analysis were performed using RaxML (Stamatakis Citation2006). Asparagopsis taxiformis (NC_031148) was used as an outgroup.
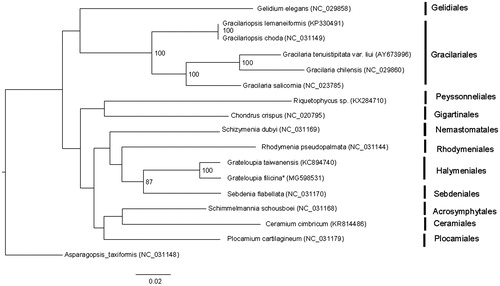
The complete plastid genome of G. filicina was a circular DNA of 195,990 bp. The overall AT content was 69.1% exhibiting a high AT richness. The plastid genome contained a set of 265 genes including 195 protein-coding genes, three rRNA genes, one tmRNA gene, 28 tRNA genes, and 38 unidentified open reading frames (ORFs). Gene arrangement and component of its plastid genome were similar to those of G. taiwanensis indicating the conserved evolution. ML analyses showed that G. filicina firstly clustered together with G. taiwanensis (). And the clustering relationship of 17 red algae supported the previous classification system. The complete plastid genome provided in this study would be useful for further understanding the evolution of Grateloupia.
Figure 1. Phylogenetic tree (maximum likelihood) of 17 representative Rhodymeniophycidae species based on rbcL and 16S genes. Numbers along branches are RaxML bootstrap supports based on 1000 nreps (<70% support not shown). Asterisks after species names indicate newly determined mitochondrial genomes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Masuda M, Kato A, Shimada S, Kawaguchi S, Phang SM. 2000. Taxonomic notes on marine algae from Malaysia. II. Seven species of Rhodophyceae. Bot Mar. 181.
- Nikapitiya C, De Zoysa M, Jeon Y-J, Lee J, Jee Y. 2007. Isolation of sulfated anticoagulant compound from fermented red seaweed Grateloupia filicina. J World Aquac Soc. 38:407–417.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxf, England). 22:2688–2690.
- Sun Y, Chen X, Cheng Z, Liu S, Yu H, Wang X, Li P. 2017. Degradation of polysaccharides from Grateloupia filicina and their antiviral activity to avian leucosis virus subgroup J. Mar Drugs. 15:345.
- Wang SC, Bligh SW, Shi SS, Wang ZT, Hu ZB, Crowder J, Branford-White C, Vella C. 2007. Structural features and anti-HIV-1 activity of novel polysaccharides from red algae Grateloupia longifolia and Grateloupia filicina. Int J Biol Macromol. 41:369–375.
- Wynne MJ. 1998. A checklist of benthic marine algae of the tropical and subtropical Western Atlantic: fourth revision. Beihefte Nova Hedwigia. 116:1–155.
- Yu Q, Yan J, Wang S, Ji L, Ding K, Vella C, Wang Z, Hu Z. 2012. Antiangiogenic effects of GFP08, an agaran-type polysaccharide isolated from Grateloupia filicina. Glycobiology. 22:1343–1352.
