Abstract
The complete mitochondrial genome of Eurasian lynx (Lynx lynx) from China has been described in this study. It has a circular genome of 16,996 bp with a higher A + T content of 58.65%, and the base composition is A: 32.31%, G: 14.29%, T: 26.35%, C: 27.06%. When compared with the reported individual (KR919624) from China, there are 81 variations and 58 base deletion between the two sequences. The phylogenetic analysis indicated our sequence separated clearly from Eurasian lynx individuals in the previous publications. These results could provide more molecular information for the conservation of Eurasian lynx genetic resources.
Eurasian lynx (Lynx lynx), a medium-sized carnivore of the family Felidae, has a wide distribution in the world. In China, the distribution of Eurasian lynx stretches from Xinjiang, Qinghai, through the southwest of China down into the northeast of China. Due to over hunting, habitat destruction, and fragmentation, Eurasian lynx is in a very low density, and thus classified as national second class protected animal in China, it is also on the list of Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) (Ablimit et al. Citation1998; Tian et al. Citation2002; Wang and Xie Citation2008). Present study situation on genetic diversity of Eurasian lynx is mainly concentrated in Europe (Hellborg et al. Citation2002; Spong and Hellborg Citation2002; Schmidt et al. Citation2011; Cömert et al. Citation2018). For the Chinese population, only one complete mitochondrial genome (mitogenome) was reported (Ning et al. Citation2016). Here, we obtained a muscle sample of Chinese Eurasian lynx from the forest public security of Tianquan county (N 30°3′59″, E 102°46′10″), China. The sample was stored at zoology laboratory, Sichuan Agricultural University (Accession: sicau -lz-A232). In this study, the complete mitogenome of Eurasian lynx was sequenced and characterized, and it could provide more molecular information for the conservation of Eurasian lynx genetic resources.
Total genomic DNA was extracted with the Phenol-chloroform method (Sambrook et al. Citation2001). Using reported Eurasian lynx (KR919624) mitogenome sequence as a reference, we designed twenty pairs of primer and amplified Eurasian lynx mitogenome successfully.
The mitogenome of Eurasian lynx has a circular genome of 16,996 bp (GenBank accession number MH 706704), including 13protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and a control region (D-loop) which was similar to typical mammalian mitochondrial DNA. The start codon of most PCGs is ATG or ATA while ND2 start with ATC. For the stop codon, seven genes ended with TAA, four ended with single T, while ND1 ended with TA, and the cytochrome b gene ended with AGA. The base composition of Eurasian lynx is A: 32.31%, G: 14.29%, T:26.35%, and C: 27.06%.AT and GC contents of mitogenome are 58.65% and 41.35%, respectively. Remarkably, when compared with the reported sequence (KR919624) from China, there are 81 variations and 58 base deletion between the two sequences, sequence similarity is 99%. The D-Loop is the hypervariable region in mitogenome, which contains 27 variations and total 58 base pairs deletion. In detail, sequence similarity is 94.5%, 58 base pairs deletion are separated in 8 sites, the longest deletion is 28 bp, exhibiting a certain degree of disparity.
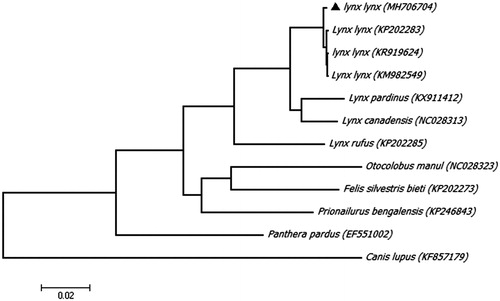
To further explore the taxonomic status of Eurasian lynx, a Maximum likelihood (ML) phylogenetic tree was reconstructed in MEGA7, using an alignment comprising 11 complete mitogenome sequences from the order Carnivora, and a grey wolf (Canis lupus) as an outgroup (). On the phylogenetic tree, all sequences from genus Lynx are clustered as a clade, but our Eurasian lynx is distinct from other Eurasian lynxes clearly, implying a different maternal origin.
Figure 1. Maximunm likelihood (ML) phylogenetic tree of Eurasian lynx (Lynx lynx) and the other 11 sequences from the order Carnivora with a sequence of grey wolf (Canis lupus) as an outgroup. Number at each node indicates the bootstrap support values. GenBank accession numbers are given in the brackets after the species name. The sequence characterized in this study is annotated with a black triangle symbol.
Acknowledgements
Our sincere thanks are given to Pu Zhao, Mengmeng Dong, Qian Su and Xue Liu for their assistance in the laboratory, and to Yuting Liu for her assistance in sample collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ablimit A, Hu DF, Ai M. 1998. Study on the ecology, distribution, resource and protection strategies of Lynx lynx in Xinjiang. Arid Zone Res. 15:38–43.
- Cömert N, Carlı O, Dinçtürk HB. 2018. The missing lynx of Eurasia at its Southern edge: a connection to the critically endangered Balkan lynx. Mitochondrial DNA A. 1–7. doi:10.1080/24701394.2018.1445240.
- Hellborg L, Walker CW, Rueness EK, Stacy JE, Kojola I, Valdmann H, Vilà C, Zimmermann B, Jakobsen KS, Ellegren H. 2002. Differentiation and levels of genetic variation in northern European lynx (Lynx lynx) populations revealed by microsatellites and mitochondrial DNA analysis. Conserv Genet. 3:97–111.
- Ning Y, Liu H, Jiang GS, Ma JZ. 2016. Phylogenetic relationship of Eurasian lynx (Lynx lynx) revealed by complete mitochondrial genome. Mitochondrial DNA. 27:3477–3478.
- Sambrook J, Russell D, Maccallum P. 2001. Molecular cloning: a laboratory manual, 3rd edition. Immunology. 49:895–909.
- Schmidt K, Ratkiewicz M, Konopiński MK. 2011. The importance of genetic variability and population differentiation in the Eurasian lynx Lynx lynx for conservation, in the context of habitat and climate change. Mammal Review. 41:112–124.
- Spong G, Hellborg L. 2002. A near-extinction event in Lynx: do microsatellite data tell the tale? Conserv Ecol. 6:289–291.
- Tian JL, Li GC, Sun HY. 2002. Current status and protection of feline lynx population in northeast China. Special Wild Economic Animal & Plant Res. 59–61.
- Wang S, Xie Y. 2008. China species red list. Beijing (BJ): Higher Education Press.
