Abstract
In this study, 45 complete mitochondrial genomes of Sciaenidae species obtained from GenBank were analyzed. Our results suggested that mitochondrial genomes could be an effective marker for resolving the phylogeny of Sciaenidae species. Phylogenetic relationships indicated that Johnius species are more ancient species among all the species that were used in this analysis because Johnius species stands on the root of the polygenetic tree. However, Collichthys niveatus could not separate from Larimichthys polyactis based on the complete mtDNA which indicate that these two species needs further study in the future.
Keywords:
The family of Sciaenidae is commercially important species which is one of the most species of the order Perciformes, with about 70 genera and more than 270 species worldwide distribution (Nelson et al. Citation2016). Sciaenidae species are very similar in morphology and synonyms always recorded in different regions [such as Argyrosomus japonicus and A. amoyensis were recorded as Nibea japonica and N. miichthioides, respectively by Zhu et al. (Citation1963)]. However, A. japonica and N. miichthioides were recorded as A. ponicus and A. amoyensis respectively by the fish database of Taiwan (fishdb.sinica.edu.tw), as well as Fishbase (fishbase.org). In addition, few studies have assessed the phylogenic relationship of the sciadaenids, and those that are available do not provide a clear explanation of the relationship among the evaluated taxon.
In this study, 25 Sciaenidae species of 45 complete mtDNA sequences were assessed from GenBank (Supplement Table 1) and were used to analyze their phylogenetic. A 600–900bp tandem repeat sequences were identified between tRNA-Thr and tRNA-Pro among the Johnius species. The Maximum Likelihood tree successfully divided the Sciaenidae species and Thunnus species, and the most all gene species were well determined except for the N. coibor, Chrysochir aureus, Collichthys niveatus, and N. miichthioides. The results of the phylogenetic tree indicated that Johnius species is a more ancient species since it stands at the root of the tree (). In addition, a 600–900bp tandem repeat sequences were identified among Johnius species making it much larger (Supplement Table 2) than other species in this study.
Figure 1. Phylogenic relationship among 48 Sciaenidae sequences from GenBank based on Maximum Likelihood method.
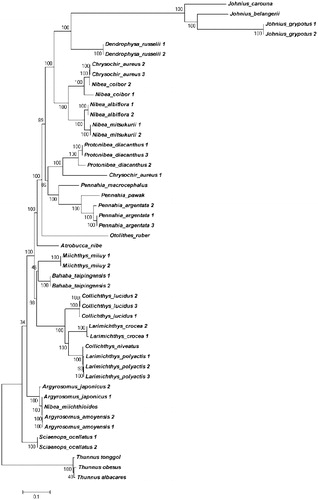
Nibea species were well claded into a separate group except for N. miichthioides, while N. miichthioides were divided into groups of A. rgyrosomus. N. miichthioides, and A. amoyensis were synonyms determined with otoliths and bladder by Zhu et al. (Citation1963) suggesting that N. miichthioides and A. amoyensiswere the same species. However, complete mtDNA failed to differentiate A. amoyensis and A. japonicus, these results were in concordance with the study done by Lv (Citation2017), stating that geographical variation may contribute to this confusion.
Chrysochir aureus 1 was divided into the Protonibea species, while C. aureus 2 and C. aureus 3 were claded with Nibea species. COI, Cytb, and ND2 were used to determine the phylogenetic relationship among Sciaenidae species, misidentification of morphological species may lead to confusing results (Lv Citation2017), yet, it may contribute to this strange phenomenon.
Collichthys lucidus were well identified out of Larimichthys crocea and L. polyactis, but, Col. niveatus could not separate from L. polyactis. Bioinformatics was used to analyze the phylogenetic relationship among eight Sciaenidae species as elaborated by Sun et al. (Citation2017). Different process results indicate that Col. niveatus was very close to L. crocea and L. polyactis, but much farther from C. lucidus. The results produced in this study were supported by analysis done by Sun et al. (Citation2017), which indicated that the taxa status of Col. niveatus needed more advanced study in the nearest future.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Lv JL. 2017. DNA barcoding and molecular phylogeny of Sciaenidae in coastal waters of China. Guangzhou: Jinan University. (In Chinese press)
- Nelson JS, Grande T, Wilson MVH. 2016. Fishes of the world. 5th ed. New York: John Wiley and Sons; p. 1–752.
- Sun YL, Yang TY, Meng W, Yang BQ, Zhang T. 2017. Analysis of the mitochondrial genome characteristics and phylogenetic relationships of eight sciaenid fishes. Mar Sci. 41:48–54. (In Chinese press)
- Zhu YD, Luo YL, Wu HL. 1963. Taxonomy system of Chinese Sciaenidae species and new species described. Shanghai: Shanghai Science and Technology press; p. 1–100. (in Chinese press)
