Abstract
The mango leafhopper, Amritodus atkinsoni is a serious and an endemic insect pest of mango in India. We analyzed mtCOI gene sequences from six Indian populations of A. atkinsoni for genetic diversity and population structure. The analysis of mtCOI sequence revealed 14 unique haplotypes and a low level of nucleotide diversity. mtCOI gene sequence analysis also revealed that A. atkinsoni specimens were clearly differentiated from other closely related species with a high level of accuracy.
Introduction
Mango leafhopper Amritodus atkinsoni (Lethierry) is one of the most economically important insect pests of mango (Mangifera indica L.) distributed mainly in South Asian and South East Asian countries (Waterhouse Citation1993). Amritodus atkinsoni is considered as the native of India and Pakistan (Pena et al. Citation1998) and causes 20–100% yield loss (Choudhary et al. Citation2012). Inspite the status of the pest, information on the population genetics of A. atkinsoni is not available from any region of the world.
Mitochondrial COI gene has been extensively used in population studies and identification of insects have unique features i.e. non recombinant, high copy numbers, simple maternal inheritance, high rate of evolution and robust evolutionary markers for determining intra- and inter-specific relationships of various invertebrate taxa including many insect species (Lunt et al. Citation1996; Armstrong and Ball Citation2005; Choudhary et al. Citation2018).
Therefore, in this study, we have employed the mitochondrial COI gene to infer the genetic diversity of A. atkinsoni collected from different regions of India. The dataset of mtCOI gene developed for the species has the potential for rapid and accurate identification.
Materials and methods
Adult A. atkinsoni were collected from six locations within India ( and ). The collected adults were identified on the basis of morphological descriptions given by Viraktamath and Mohan (Citation2004). Voucher specimens were preserved with voucher ID: A1, A2, B1, B2, B3, B4 P3, P5, P8 and P6 in the Entomology Laboratory, ICAR RCER, Research Centre, Ranchi, India.
Figure 1. (A) Sites of Amritodus atkinsoni collections for the present study. BH: Bihar, JH: Jharkhand, OD: Odisha, TL: Telangana, GU: Gujarat and RJ: Rajasthan. (B) Median joining network of mtCOI haplotypes of Amritodus atkinsoni. Each circle represents haplotype, and size of the circle is proportional to the haplotype frequency. Colours indicate the proportion of individuals of different populations present in a haplotype. (C) Neighbor-joining tree of 102 mtCOI gene sequences of different species representing family Cicadellidae. Bootstrap values based on 1000 replications are shown near nodes.
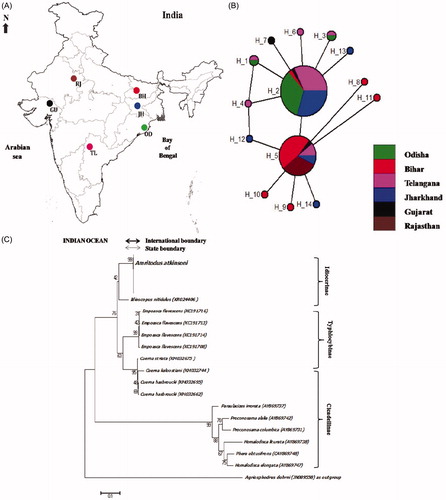
Table 1. Details of sampling locations and genetic diversity indices of Amritodus atkinsoni.
Total genomic DNA was extracted from individual specimens with methods described in Prabhakar et al. (Citation2009). The mtCOI gene was amplified using UEA7 (F) and UEA10 (R) developed by Lunt et al. (Citation1996) with PCR conditions mentioned in Prabhakar et al. (Citation2012). These were then freeze-dried and custom sequenced using same upstream and downstream primers (Xcelris Labs Limited, India).
GenBank Accession Numbers: KC513467- KC513474 and KY084430-KY084442.
Nucleotide sequence analysis
The 85 mtCOI gene sequences of A. atkinsoni were aligned using ClustalW implemented in MEGA 6.0. (Tamura et al. Citation2013). Descriptive statistics of sequences were calculated with DnaSP (Librado and Rozas Citation2009). A median-joining network was constructed using NETWORK 4.6 (Bandelt et al. Citation1999). Phylogenetic reconstruction with 102 sequences representing the family Cicadellidae were analysed with the neighbor-joining method in MEGA 6.0. The “best close match” method in the programme TaxonDNA (Meier et al. Citation2006) was performed to test the frequency of successful identification.
Results and discussion
mtCOI gene sequences of 537 bp were obtained and aligned from 85 individuals of A. atkinsoni from six populations in India. The number of haplotypes per population (H) ranged from 1 to 6, haplotype diversity (Hd) from 0 to 1.000, average number of nucleotide differences within populations (K) from 0 to 0.23529 ± 0.28567 and nucleotide diversity (π) ranged from 0 to 0.00248 ± 0.002551 (). A total of 14 haplotypes were identified. The most common haplotype was H2 and H5 comprised of 43 and 28 individuals of A. atkinsoni, respectively. Haplotype 2 (H2) was shared by five populations [JH (Jharkhand), GU (Gujarat), TL (Telangana), BH (Bihar) and OD (Odisha)] except RJ (Rajasthan) (). The analysis of mtCOI sequences demonstrated low values of nucleotide diversity and relatively high value of haplotype diversity with no geographic pattern. Most of the populations shared one or both of the most frequent haplotypes (H2 and H5). When values of haplotype (Hd > 0.5) and nucleotide (π > 0.5%) diversity are high, the population appears to be stable with a relatively long evolutionary history (Rosetti and Remis Citation2012). None of the population presently examined meet these conditions, suggesting recent and unstable evolutionary history of the species. Low values of π and high values of Hd suggest the existence of small populations that have suffered recent population growth. Except R, this situation is observed for all the populations studied, where haplotype diversity is relatively high and nucleotide diversity relatively low. When both π and Hd values are low, the population has probably undergone a reduction in population size or a recent colonization event (Rosetti and Remis Citation2012). RJ is characterized by low values in both indices, reflecting recent population colonization.
The mtCOI gene analysis in the present study reveals A. atkinsoni to exhibit a high degree of diversity when compared with other hopper species. In Scaphoideus titanus Ball, COII gene sequences with 22 European and 8 Asian populations showed Hd and (π) value of 0.583 ± 0.072 & 0.203 ± 0.042 and 0.0046 ± 0.0027 & 0.0013 ± 0.0001 for nucleotide diversity, respectively, and showed moderate values for haplotype and nucleotide diversity (Papura et al. Citation2012). Genetic analysis of mtCOI gene sequences among 11 populations of rice brown planthopper, Nilaparvata lugens (Stål) and 5 populations of whitebacked planthopper, Sogatella furcifera (Horvath) from southeast Asia, showed low values of haplotype diversity 0.000–0.299 and 0.188–0.382 for N. lugens and S. furcifera, respectively (Mun et al. Citation1999), that were substantially lower than those observed in present study.
One hundred and two sequences from the family Cicadellidae were analysed for the accurate identification of A. atkinsoni. Intraspecific genetic divergence of 85 mtCOI gene sequences of A. atkinsoni ranged between 0 and 0.9% with an average of 0.50%. The best close match methods identified the 95% intraspecific genetic divergence threshold value 0.44% of A. atkinsoni identified all A. atkinsoni specimens correctly. Our results showed large gaps between intra and interspecific genetic distances for cicadellid species are consistent with previous reports on mtCOI gene based identification ofof insect species (Choudhary et al. Citation2018). DNA species barcode gaps can be detected by recording the overlap between the highest intraspecific and the lowest interspecific genetic distances (Meier et al. Citation2008). Phylogenetic analysis using the neighbor-joining method () gave a strong bootstrap support (99%) for the monophyly of A. atkinsoni. The mtCOI gene sequences developed in the present study and its availability in GenBank could also be useful in identification of the species in the future (Virgilio et al. Citation2010; Jinbo et al. Citation2011). Incorrect species identification using the mtCOI is largely down to insufficient geographic sampling of the taxa (Jinbo et al. Citation2011). In this context and in relation to A. atkinsoni, our present results significantly increase the number of mtCOI gene sequences available, which will encourage the molecular identification of this and related important mango leafhopper species.
Acknowledgements
Authors are grateful to Mr. Deepak Tanti and Mr. Pradeep Lakra for collection of mango leafhoppers.
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Armstrong KF, Ball SL. 2005. DNA barcodes for biosecurity: invasive species identification. Philos Trans R Soc Lond B. 360:1813–1823.
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48.
- Choudhary JS, Naaz N, Lemtur M, Das B, Singh AK, Bhatt BP, Prabhakar CS. 2018. Genetic analysis of Bactrocera zonata (Diptera: Tephritidae) populations from India based on cox1 and nad1 gene sequences. Mitochondr DNA A. 29:727–736. DOI: 10.1080/24701394.2017.1350952
- Choudhary JS, Prabhakar CS, Maurya S, Kumar R, Das B, Kumar S. 2012. New report of Hirsutella sp. infecting mango hopper Idioscopus clypealis from Chotanagpur Plateau, India. Phytoparasitica. 40:243–245.
- Jinbo U, Kato T, Ito M. 2011. Current progress in DNA barcoding and future implications for entomology. Entomol Sci. 14:107–124.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Lunt DH, Zhang DX, Szymura JM, Hewitt GM. 1996. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 5:153–165.
- Meier R, Shiyang K, Vaidya G, Ng PKL. 2006. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 55:715–728.
- Meier R, Zhang G, Ali F. 2008. The use of mean instead of smallest interspecific distances exaggerates the size of the "barcoding gap" and leads to misidentification. Syst Biol. 57:809–813.
- Mun J, Song Y, Heong KL, Roderick GK. 1999. Genetic variation among Asian populations of rice planthoppers, Nilaparvata lugens and Sogatella furcifera (Hemiptera: Delphacidae): mitochondrial DNA sequences. Bull Entomol Res. 89:245–253.
- Papura D, Burban C, van Helden M, Giresse X, Nusillard B, Guillemaud T, Kerdelhué C. 2012. Microsatellite and mitochondrial data provide evidence for a single major introduction for the Neartic Leafhopper Scaphoideus titanus in Europe. PLoS One. 7:e36882.
- Pena JE, Mohyuddin AI, Wysoki M. 1998. A review of the pest management situation in mango agroecosystems. Phytoparasitica. 26:129–148.
- Prabhakar CS, Mehta PK, Sood P, Singh SK, Sharma P, Sharma PN. 2012. Population genetic structure of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae) based on mitochondrial cytochrome oxidase (COI) gene sequences. Genetica. 140:83–91.
- Prabhakar CS, Sood P, Kapoor V, Kanwar SS, Mehta PK, Sharma PN. 2009. Molecular and biochemical characterization of three bacterial symbionts of fruit fly, Bactrocera tau (Tephritidae: Diptera). J Gen App Microbiol. 55:213–220.
- Rosetti N, Remis MI. 2012. Spatial Genetic Structure and Mitochondrial DNA Phylogeography of Argentinean Populations of the Grasshopper Dichroplus elongatus. PLoS one. 7:e40807.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Viraktamath CA, Mohan GS. 2004. A revision of the Idiocerine leafhopper genus Amritodus (Hemiptera: Cicadellidae) breeding on mango. Entomon. 578:1–117.
- Virgilio M, Backeljau T, Nevado B, Meyer MD. 2010. Comparative performances of DNA barcoding across insect orders. BMC Bioinformatics. 11:206.
- Waterhouse DF. 1993. The major arthropods pests and weeds of agriculture in South-East Asia; Distribution, Importance and Origin. Monograph. No. 21.
