Abstract
The mitochondrial genome plays an important role in studies on phylogeography and population genetic diversity. Here we report the complete mitochondrial genome of Lupocycloporus gracilimanus (Stimpson, 1858) which is the first mitochondrial genome reported in genus Lupocycloporus by now. The mitogenome is 15,990 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and a putative control region. The phylogenetic analysis showed that L. gracilimanus was closest to genus Scylla. The present research should provide valuable information for phylogenetic analysis and classification of Portunidae.
The small hand swimming crab, Lupocycloporus gracilimanus (Stimpson, 1858), is under Portunidae. It is mainly distributed in coasts of China, Australia, New Zealand, Philippines, Malaysia, Andaman (Dai et al. Citation1986), and also recorded along the west coast of the Indian Ocean in 2005 (Dineshbabu Citation2005). Additionally, L. gracilimanus is one of the dominant crab species in autumn in East China Sea (Yu et al. Citation2005). So far, limited information about the phylogeny and genetic diversity are available. To well-understand the evolutionary status of L. gracilimanus and their position in family Portunidae, we amplified and sequenced the complete mitochondrial genome of L. gracilimanus.
Specimens of L. gracilimanus were collected from Weizhou Island (21.0234N, 109.0940E), Guangxi province of China, and deposited at the Marine Biology Institute of Shantou University, Shantou of China. Genomic DNA was extracted from muscle tissue using traditional phenol–chloroform method. Long and conventional PCR methods were applied to obtain the whole mitogenome sequence. The mitochondrial genome size of L. gracilimanus is 15,990 bp (GenBank accession number: MH729187), including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and a putative control region. The composition and order of genes were consistent with other species such as Portunus (Yamauchi et al. Citation2003; Ma et al. Citation2013) and Charybdis (Ma et al. Citation2015; Yang et al. Citation2017). The overall mitogenome composition is 34.05% for A, 11.23% for G, 19.02% for C and 35.70% for T, respectively. Among all the 37 genes, 23 were encoded by heavy strand and 14 were encoded by light strand. Three types of initiation codons were found in the 13 protein-coding genes, including ATG (COX1, COX2, ATP8, COX3, ND5, ND4, ND4L, ND6, Cytb and ND2 gene), ATT (ATP6 and ND3 gene) and ATA (ND1 gene). Four kinds of termination codons including two incomplete termination codons (T-, TAG, TA- and TAA) were also found.
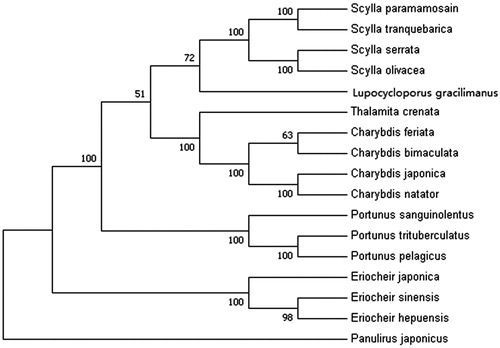
In order to analyze the phylogenetic position of L. gracilimanus, 12 protein-coding genes (except ND6) from 16 crab species in GenBank database were used to construct the phylogenetic tree by maximum-likelihood (ML) method (). Panulirus japonicus was served as an outgroup for tree rooting. The result showed that L. gracilimanus was genetically closer to genus Scylla (S. paramamosain, S. tranquebarica, S. serrata and S. olivacea) than to genus Portunus (P. sanguinolentus, P. trituberculatus and P. pelagicus). This is consistent with the previous research by Evans (Citation2018). Moreover, Lupocycloporus was recommended as a genus in subfamily Lupocyclinae according to their morphological similarity (Spiridonov et al. Citation2014). This study should provide valuable data for phylogenetic analysis and classification for Portunidae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dai A, Yang S, Song Y, Chen G. 1986. Marine crabs in China. Beijing: China Ocean Press.
- Dineshbabu AP. 2005. First record of hairy crabs, Portunus (Monomia) gracilimanus along west coast of India. Mar Fish Infor Serv, T & E Ser. 184:16–17.
- Evans N. 2018. Molecular phylogenetics of swimming crabs (Portunoidea Rafinesque, 1815) supports a revised family-level classification and suggests a single derived origin of symbiotic taxa. PeerJ. 6:e4260.
- Ma H, Ma C, Li C, Lu J, Zou X, Gong Y, Wang W, Chen W, Ma L, Xia L. 2015. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and population genetics. Sci Rep. 5:11524.
- Ma H, Ma C, Li X, Xu Z, Feng N, Ma L. 2013. The complete mitochondrial genome sequence and gene organization of the mud crab (Scylla paramamosain) with phylogenetic consideration. Gene. 519:120–127.
- Spiridonov VA, Neretina TV, Schepetov D. 2014. Morphological characterization and molecular phylogeny of Portunoidea Rafinesque, 1815 (Crustacea Brachyura): implications for understanding evolution of swimming capacity and revision of the family-level classification. Zool Anz. 253:404–429.
- Yamauchi MM, Miya MU, Nishida M. 2003. Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene. 311:129–135.
- Yang X, Ma H, Waiho K, Fazhan H, Wang S, Wu Q, Shi X, You C, Lu J. 2017. The complete mitochondrial genome of the swimming crab Chaybdis natator (Herbst) (Decapoda: Brachyura: Portunidae) and its phylogeny. Mitochondrial DNA B. 2:530–531.
- Yu C, Song H, Yao G. 2005. Crab community structure in the East China Sea. Oceanol Limnol Sin. 36:213–220.