Abstract
Clinacanthus nutans (Burm.f.) Lindau. is a medicinal species occurring in southern Guangdong province, Guangxi province, Hainan province and Yunnan province of China, which has been widely used in the medical field. However, its complete plastome information and systematic position are still unknown. In order to enhance the development of its medicinal value, we report and characterize the complete plastid genome sequence of C. nutans in an effort to provide genomic resources useful to promoting its medicinal value and systematic research. The complete plastome is 151,669 bp in length and contains the typical structure and gene content of angiosperm plastome, including two Inverted Repeat (IR) regions of 25,366 bp, a Large Single-Copy (LSC) region of 83,502 bp and a Small Single-Copy (SSC) region of 17,435 bp. The plastome contains 113 genes, consisting of 80 unique protein-coding genes, 29 unique tRNA gene and four unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA and 16S rRNA). The overall A/T content in the plastome of C. nutans is 61.60%. The complete plastome sequence of C. nutans will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Acanthaceae.
Introduction
As a medicinal plant in family Acanthaceae, Clinacanthus nutans (Burm.f.) Lindau. is native to many tropical Asia countries including Thailand and often cultivated. C. nutans has been traditionally used as a medicine for topical treatment of skin rashes, insect and snake bite, Herpes simplex virus, and varicella-zoster virus lesions (Pongmuangmul et al. Citation2016). Clinacanthus nutans was sampled from the greenhouse within Hainan University campus, Haikou, Hainan, China. A voucher specimen (Wang et al., U2) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Andrographis paniculata (NC_022451.2) (Ding et al., Citation2013, unpublished) using MITO bim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Andrographis paniculata (NC_022451.2). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of C. nutans is found to possess a total length 151,669 bp with the typical quadripartite structure of angiosperms, contains two Inverted Repeats (IRs) of 25,366 bp, a Large Single-Copy (LSC) region of 83,502 bp and a Small Single-Copy (SSC) region of 17,435 bp. The plastome contains 113 genes, consisting of 80 unique protein-coding genes (seven of which are duplicated in the IR: rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and rps12), 29 unique tRNA genes (seven of which are duplicated in the IR) and four unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA and 16S rRNA). Among these genes, three pseudogenes [rpoC2 (translation from 20,156 to 16,003), ycf15 (translation from 142,500 to 142,436 and translation from 92,491 to 92,673), and ycf1 (translation from 125,438 to 121,695)], 15 genes possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as atypical of angiosperms. The overall A/T content in the plastome of C. nutans is 61.60%, which the corresponding value of the LSC, SSC, and IR regions were 63.50%, 67.60%, and 56.50%, respectively.
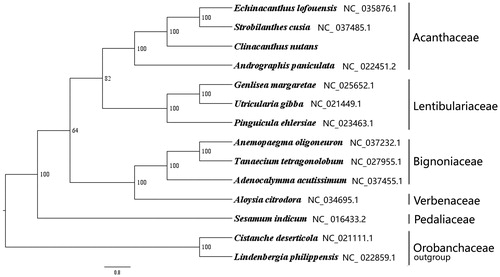
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum-likelihood (ML) phylogeny of three published complete plastomes of Acanthaceae, three published complete plastomes of Lentibulariaceae, three published complete plastomes of Bignoniaceae, one published complete plastomes of Verbenaceae, one published complete plastomes of Pedaliaceae, using Cistanche deserticola (Orobanchaceae, Lamiales) and Lindenbergia philippensis (Orobanchaceae, Lamiales) as outgroups. The phylogenetic analysis indicates that C. nutans is closer to Andrographis paniculata than other species in this study (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of C. nutans will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Acanthaceae.
Figure 1. The best ML phylogeny recovered from 14 complete plastome sequences by RAxML. Accession numbers: Clinacanthus nutans (This study, GenBank accession number: MH778102), Echinacanthus lofouensis NC_ 035876.1, Strobilanthes cusia NC_ 037485.1, Andrographis paniculata NC_ 022451.2, Utricularia gibba NC_021449.1, Genlisea margaretae NC_025652.1, Pinguicula ehlersiae NC_023463.1, Anemopaegma oligoneuron NC_037232.1, Tanaecium tetragonolobum NC_027955.1, Adenocalymma acutissimum NC_037455.1, Aloysia citrodora NC_034695.1, Sesamum indicum NC_ 016433.2, outgroups: Cistanche deserticola NC_021111.1; Lindenbergia philippensis NC_ 022859.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ding P, Shao YH, Wang JG, Wu XW, Fu XC, Huang S., Lai XP. 2013. Andrographis paniculata chloroplast complete genome. National Center for Biotechnology. Unpublished. https://www.ncbi.nlm.nih.gov/nuccore/NC_022451.2/
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Pongmuangmul S, Phumiamorn S, Sanguansermsri P, Wongkattiya N, Fraser IH, Sanguansermsri D. 2016. Anti-herpes simplex virus activities of monogalactosyl diglyceride and digalactosyl diglyceride from Clinacanthus nutans, a traditional Thai herbal medicine. Asian Pacific J Tropical Biomed. 6:192–197.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
