Abstract
In this study, the complete mitochondrial genome of Neopagetopsis ionah was obtained, which was 17,634 bp including two ribosomal RNAs, 13 protein-coding genes, 22 transfer RNAs, and a non-coding control region. In 13 protein-coding genes, there types of initiation codon (ATC, ATG, and GTG) and four types of stop codons (TAA, TAG, TA, and T) were identified. Among the 22 transfer RNAs, eight tRNAs were encoded by L-strand. The length of D-loop was 1519 bp and its contents of A, T, C, and G were 26.9%, 27.6%, 17.5%, and 30%, respectively. The complete mtDNA sequences of N. ionah and other 13 species were used to reconstruct the phylogenetic tree suggested that N. ionah was closest to some species of Channichthyidae. The study would provide a basic data for further research on population structure, conservation genetics and molecular evolution of N. ionah.
Neopagetopsis ionah (Channichthyidae, Neopagetopsis), the only known member of its genus, is a species of crocodile icefish found in the Southern Ocean (Circum-Antarctic on the continental shelf and slope) at depths of from 20 to 900 m (Gon and Heemstra Citation1990). The optimum water temperature is 1 °C. The species grows to a length of 56 cm and adults mainly prey on fishes and krill (Gon and Heemstra Citation1990). Neopagetopsis ionah is the only known member of its genus. So far, no complete mitochondrial sequence information is available. The study is important for the Antarctic ecosystem and further research on genetics and evolution of N. ionah.
Adult fish of N. ionah was collected from Antarctic (60°21′36″S, 46°41′24″W), it was transported to East China Sea Fisheries Research Institute, Chinese Academy of Fishery Science after freezing at −80 °C. The genomic DNA was extracted from muscle tissues using Animal Genomic DNA Extraction Kit (Tiangen, Beijing, Chian) following the operation manual. The amplifying and sequencing primers were designed according to the sequence of Chionodraco rastrospinosus (MF622064) and Chionodraco hamatus (KT921282) and sequenced using the Illumina HiSeq2000 platform (Illumina, San Diego, CA).
We obtained the complete mitochondrial sequence of N. ionah and submitted it into the Genbank database with an accession number MF596172. This complete mitochondrial genome is 17,634 bp in length, including 13 protein-coding genes, tw rRNA, 22 tRNA, and a control region. The overall nucleotide composition is 25.46% for A, 26.34% for T, 19.01% for C, and 29.19% for G. In 13 protein-coding genes, three kinds of start codons were identified (ATG, ATC, and GTG), ND1 and CO2 were started with GTG and ATC, respectively, whereas others started with ATG. Four types of stop codons (TAA, TAG, TA, and T) were detected. Eight genes (ND6, ND4, ND3, ND2, ND1, CO1, CO3, and Cytb) ended with T. Among the 22 tRNAs, eight tRNAs (tRNAPro, tRNAGlu, tRNASer, tRNATyr, tRNACys, tRNAAsn, tRNAAla, and tRNAGln) were encoded by L-strand, when others were encoded by H-strand. The length of D-loop was 1519 bp and its overall base composition was 26.9% for A, 27.6% for T, 17.5% for C, and 30% for G, with a high AT content of 54.5%, accord with the structural characteristics of AT rich (Song et al. Citation2016, Liang et al. Citation2018). It also has many different lengths of base repeat.
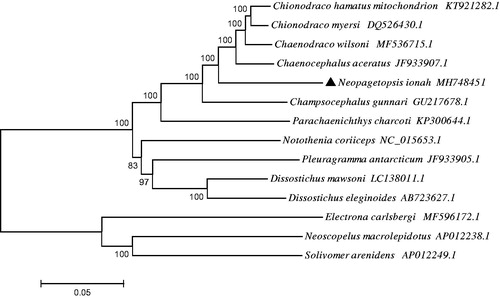
To evaluate the phylogenetic position of N. ionah, the phylogenetic tree was reconstructed based on complete mtDNA sequences using the neighbour-joining method in MEGA 5.1 (Kumar et al. Citation2008) (). Solivomer arenidens (Myctophiformes, Neoscopelidae), Neoscopelus macrolepidotus (Myctophiformes, Neoscopelidae) and Electrona carlsbergi (Myctophiformes, Myctophidae) were used as an out-group. The phylogenetic tree showed that N. ionah clustered with C. hamatus, C. rastrospinosus and other fishes in family Channichthyidae. Besides, the out-group formed a big sister branch as well.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Gon O, PC. Heemstra. 1990. Fishes of the Southern Ocean. South African Institute for Aquatic Biodiversity, the Hebrew University of Jerusalem, 462.
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinfo. 9:299–306.
- Liang SZ, Song W, Huang HL, Qu TC, Zhang FY, Jiang KJ, Chen XZ, Ma LB. 2018. The complete mitochondrial genome of Chionodraco rastrospinosus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B. 3:816–817.
- Song W, Li LZ, Huang HL, Meng YY, Jiang KJ, Zhang FY, Chen XZ, Ma LB. 2016. The complete mitochondrial genome of Chionodraco hamatus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B. 1:52–53.