Abstract
In this study, genetic diversity and population genetic structure of Sagitta enflata in the northern South China Sea were investigated by 623 bp fragment of mtDNA COI gene sequence. A total of 146 individuals were collected from nine stations and 92 different haplotypes were obtained. 485 variable sites (210 were parsimony informative and 275 were singleton variable sites), and no insertion or deletion was found. An analysis of molecular variance (AMOVA) and conventional population statistics (FST) revealed a low level of genetic differentiation among nine populations (FST = 0.14794, p < .05), indicating no geographical patterning among nine populations. The present results were able to provide a reference for the phylogenetic relationships and assessment of the genetic structure of S. enflata in the northern South China Sea.
Introduction
Chaetognaths (arrow worms) are small-sized and important predators in the marine ecosystem (Feigenbaum Citation1979), and copepods are the dominant prey of chaetognaths (Jennings et al. Citation2010). They can be found in coastal areas and in oceanic waters, from polar to tropical areas, and from the surface to several thousand meters depth (Alvarino Citation1965). The origins of the arrow worms remain obscure, but molecular studies will finally bring the true evolutionary relationship (Telford Citation2004). Taken as a whole, the genetic diversity of marine species is believed to present high levels within population and low levels between populations (Meriam et al. Citation2015). Many physical factors, such as climate, ocean currents, and lack of barriers in the open sea may explain this diversity (Maltagliati et al. Citation2002, Citation2010; Fernández et al. Citation2011).
Sagitta is a genus of holoplanktonic chaetognaths with about 70 species identified from the oceans around the world. These species numerically dominate mesozooplankton and are important secondary consumers in the pelagic ecosystem (Pierrot-Bults Citation1982). Sagitta enflata is a cosmopolitan epiplanktonic species in temperate coastal waters, tropical-subtropical epipelagic waters, and tropical-subtropical mesopelagic waters and occurs mainly in the upper 300 m (Øresland Citation2000; Tse et al. Citation2007). While several investigators have studied their wide geographic distribution and migration behavior, the evaluation of its genetic diversity and population genetic structure is also very important and has not been reported so far.
Mitochondrial DNA (mtDNA) is widely being used for elucidating molecular systematics studies. Moreover, cytochrome oxidase subunit I (COI) is considered as more rapid evolutionary rate and polymorphic than other mitochondrial genes, and therefore frequently used to study population genetic structure analysis and phylogeographic relationships of marine species (Rabaoui et al. Citation2011). In this study, we used sequences of the COI gene to investigate the genetic diversity and population variation of 146 S. enflata individual among nine populations from the different geographical distribution of northern South China Sea. The primary specimens have been deposited in the South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China, and the accession number is SCSMBC040498. The results would be helpful for phylogenetic reconstructions and population genetic structure of this species.
Materials and methods
Sample collection and DNA extraction
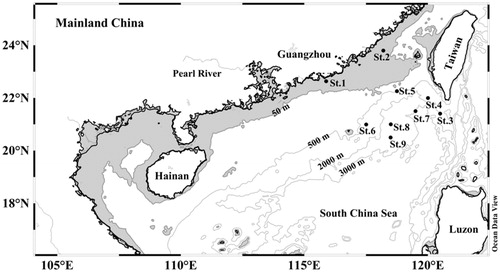
We collected 146 S. enflata individuals at nine stations in the northern South China Sea () during August 2015. Each individual was preserved in 95% ethanol before genomic DNA (gDNA) extraction. Total gDNA was extracted using marine animals gDNA kit (Biomiga, GD3311-02) and was visualized on 1.0% agarose gel to verify the quality of high molecular weight DNA extractions. Fragments of mitochondrial COI from all individuals of S. enflata were amplified using the primer pairs LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198(5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al. Citation1994). Amplifications were performed in a total volume of 50 μL containing 10 × PCR buffer, 2.5 mM dNTP Mix, 10 μM each primer, 100 ng templates, and 2 U Taq DNA polymerase (Takara, Dalian, China). The PCR program was carried out under the following conditions: an initial denaturing at 94 °C for 4 min, followed by 30 cycles of denaturing at 94°C for 30 s, annealing at 50 °C for 50 s, and extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. The PCR products were visualized on 1% agarose gels, and purified with a Takara Agarose Gel DNA Purification Kit (Takara, China). Gene sequencing was performed on ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA).
Data analyses
Both sequences (forward and reverse) of the COI gene fragment obtained for each specimen were aligned and edited by visual inspection using SeqScape version 2.5 (Applied Biosystems). Final alignments were optimized using BioEdit version 7.0.4.1 using S. enflata COI sequences as reference (GenBank accession no. KF977332). Nucleotide composition and variable sites were analyzed using MEGA3.1 (Kumar et al. Citation2004). Population structure of S. enflata was investigated using analysis of molecular variance (AMOVA), and the FST was examined using the Mantel test with 1000 permutations, performed by Arlequin version 3.01 (Excoffier et al. Citation2007).
Results and discussion
Molecular genetic studies have shown the existence of genetic differentiation corresponding to different water masses or biogeographic boundaries in several zooplankton species (Bucklin et al. Citation2000; Goetze Citation2005). The investigations on chaetognaths were performed on some widespread species like Parasagitta elegans, Parasagitta setosa, Caecosagitta macrocephala, Eukrohnia hamata (Peijnenburg et al. Citation2005, Citation2006; Miyamoto et al. Citation2010; Kulagin et al. Citation2014). In this study, amplification of a 623bp fragment of the COI gene from 146 S. enflata individuals yielded 92 distinct haplotypes (GenBank accession nos. KX009784–KX009875). Among all 92 haplotypes, there were 485 variable sites (210 were parsimony informative and 275 were singleton variable sites), and no insertion or deletion was found. The mean total nucleotide composition was 22.4% A, 19.3% G, 23.0% C, and 35.3% T.
Compared to the previous reported studies on zooplankton, our estimates of genetic diversity within the S. enflata species are close to estimates of genetic diversity within species from other taxa. The analysis of molecular variance (AMOVA) is one of the most widely used methods of genetic data analysis (Excoffier et al. Citation1992). AMOVA indicated that 15.69% of variation was attributed to distribution among populations within groups and 85.21% to the distribution within populations (). Our results showed that the genetic diversity of S. enflata for the present study region included continental shelf, continental slope, and deep-sea basins, it may be explained by the Kuroshio current intrusion through the Luzon Strait into the nSCS. In order to further study the genetic structure of this worldwide species, more molecular markers and populations will be needed in a comprehensive analysis.
Table 1. AMOVA analysis of mtDNA COI gene sequences in nine populations of S. enflata.
Acknowledgments
We are grateful to the captain, crew, and scientists of R/V Shiyan 3 for their efforts in collecting the zooplankton samples. Special thanks are due to Dr. Kaizhi Li of South China Sea Institute of Oceanology for her help in coordinating the sampling and relevant project.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alvarino A. 1965. Chaetognaths. Oceanogr Mar Biol Annu Rev. 3:115–194.
- Bucklin A, Astthorsson OS, Gislason A, Allen LD, Smolenack SB, Wiebe PH. 2000. Population genetic variation of Calanus finmarchicus in Icelandic waters: preliminary evidence of genetic differences between Atlantic and Arctic populations. ICES J Mar Sci. 57:1592–1604.
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 131:479–491.
- Excoffier L, Laval G, Schneider S. 2007. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 1:47–50.
- Feigenbaum D. 1979. Daily ration and specific daily ration of the chaetognath Sagitta enflata. Mar Biol. 54:75–82.
- Fernández MV, Heras S, Maltagliati F, Turco A, Roldán MI. 2011. Genetic structure in the blue and red shrimp, Aristeus antennatus and the role played by hydrographical and oceanographical barriers. Mar Ecol Prog Ser. 421:163–171.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3:294–299.
- Goetze E. 2005. Global population genetic structure and biogeography of the oceanic copepods Eucalanus hyalinus and E. spinifer. Evolution. 59:2378–2398.
- Jennings RM, Bucklin A, Pierrot-Bults A. 2010. Barcoding of arrow worms (Phylum Chaetognatha) from three oceans: genetic diversity and evolution within an enigmatic phylum. PLoS One. 5:e9949.
- Kulagin DN, Stupnikova AN, Neretina TV, Mugue NS. 2014. Spatial genetic heterogeneity of the cosmopolitan chaetognath Eukrohnia hamata (Möbius, 1875) revealed by mitochondrial DNA. Hydrobiologia. 721:197–207.
- Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics. 5:150–163.
- Maltagliati F, Belcari P, Casu D, Casu M, Sartor P, Vargiu G, Castelli A. 2002. Allozyme genetic variability and gene flow in Octopus vulgaris (Cephalopoda, Octopodidae) from the Mediterranean Sea. Bull Mar Sci. 71:473–486.
- Maltagliati F, Giuseppe GD, Barbieri M, Castelli A, Dini F. 2010. Phylogeography and genetic structure of the edible sea urchin Paracentrotus lividus (Echinodermata, Echinoidea) inferred from the mitochondrial cytochrome b gene. Biol J Linn Soc. 100:910–923.
- Meriam T, Wafa T, Khawla T, Tarek H, Abdeljelil G, Mhamed E. 2015. Genetic diversity and population structure of Sepia officinalis from the Tunisian cost revealed by mitochondrial COI sequences. Mol Biol Rep. 42:77–86.
- Miyamoto H, Machida RJ, Nishida S. 2010. Genetic diversity and cryptic speciation of the deep sea chaetognath Caecosagitta macrocephala (Fowler, 1904). Deep-Sea Res Part II. 57:2211–2219.
- Øresland V. 2000. Diel feeding of the chaetognath Sagitta enflata in the Zanzibar Channel, western Indian Ocean. Mar Ecol Progress. 193:117–123.
- Peijnenburg K, Haastrecht EKV, Fauvelot C. 2005. Present day genetic composition suggests contrasting demographic histories of two dominant chaetognaths of the North-East Atlantic, Sagitta elegans and S. setosa. Mar Biol. 147:1279–1289.
- Peijnenburg KTCA, Fauvelot C, Breeuwer JAJ, Menken SBJ. 2006. Spatial and temporal genetic structure of the planktonic Sagitta setosa (Chaetognatha) in European seas as revealed by mitochondrial and nuclear DNA markers. Mol Ecol. 15:3319–3338.
- Pierrot-Bults AC. 1982. Vertical distribution of Chaetognatha in the Central Northwest Atlantic near Bermuda. Biol Oceanogr. 2:31–61.
- Rabaoui L, Mejri R, Tlig-Zouari S, Bahri L, Hassine OKB, Tsigenopoulos CS. 2011. Genetic variation among populations of the endangered fan mussel Pinna nobilis (Mollusca: Bivalvia) along the Tunisian Coastline. Hydrobiologia. 678:99–111.
- Telford MJ. 2004. Evolution: affinity for arrow worms. Nature. 431:254–256.
- Tse P, Hui SY, Wong CK. 2007. Species composition and seasonal abundance of Chaetognatha in the subtropical coastal waters of Hong Kong. Estuarine Coastal Shelf Sci. 73:290–298.