Abstract
Desarmillaria tabescens is one of the most important edible, medicinal, and phytopathogenic basidiomycetes. The complete mitochondrial genome of this species was determined using next-generation sequencing technology. This mitogenome is a circular molecule of 93,439 bp with a GC content of 29.28% and contains 15 protein-coding, two rRNA (rnl and rns), and 24 tRNA genes. Phylogenetic analysis revealed that D. tabescens is genetically closest to Agrocybe aegerita. Desarmillaria tabescens mitogenome can contribute to our understanding of the phylogeny and evolution of this species.
Desarmillaria tabescens was classified in the genus Armillaria but has been recently reassigned to genus Desarmillaria based on phylogenetic analysis of 6 genetic loci (28S nuclear ribosomal large subunit DNA, Elongation Factor 1-α [EF1α], RNA polymerase II [RPB2], Actin-1 [actin-1], Glyceraldehyde-3-Phosphate Dehydrogenase [gpd], and Beta-Tubulin [TUB] and phenotypes such as exannulate mushroom formation and rhizomorph development under natural conditions (Koch et al. Citation2017). Desarmillaria tabescens is an edible mushroom (Sterry and Hughes Citation2009) and is considered a potent hepatoprotective remedy (Lu et al. Citation2007). Under natural conditions, this mushroom causes armillaria root rot (Amiri et al. Citation2008; Baumgartner et al. Citation2018) and participates in a symbiotic relationship with Galeola septentrionalis (Terashita and Chyuman Citation1987). Studies related to D. tabescens had been primarily limited to its phylogenetic relationships and distribution (Hasegawa et al. Citation2011; Coetzee et al. Citation2015; Park et al. Citation2017) and phytopathology (Amiri et al. Citation2008; Baumgartner et al. Citation2018). This study is the first to report the complete mitogenome sequence of D. tabescens (GenBank accession no. MH823225) and the phylogenetic analysis of this mushroom in relation to other species.
The A014 strain of D. tabescens used in this study was isolated from a fruit body of Quercus acutissima collected at Chungbuk National University (Cheongju, Chungcheongbuk-do, Korea; N36°37′43.64″, E127°27′04.87″). The strain has been deposited and maintained at the Korean Agricultural Culture Collection (Wanju, Jeollabuk-do, Korea) under the accession number KACC 54704. Total genomic DNA was extracted from mycelia cultured in malt extract broth media for 10 days under dark conditions using a GenEX Plant Kit (Geneall, Seoul, Korea) following the manufacturer’s instructions and sequenced using an Illumina Miseq Platform. Illumina paired-end reads were implemented de novo assembly after filtered the high-quality reads (>20 phred) using CLC de novo assembler (version 4.2.1, https://www.qiagenbioinformatics.com/products/clc-assembly-cell/; Kim et al. Citation2015). In brief, the mitochondrial contigs were compared with Flammulina velutipes mitochondrion sequence (GenBank accession JF799107) as reference by NUCmer tool in MUMmer package. Then merger of selected mitochondrial contigs were by read mapping using CLC read mapper. The genes in the mitogenome were annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html; Tillich et al. Citation2017), and manual confirmed using Artemis annotation tool was based on BLAST searches.
The complete mitogenome of strain A014 of D. tabescens is a circular molecule of 93,439 bp with a GC content of 29.28%. This mitogenome contains 15 protein-coding, 24 tRNA, and two rRNA (rnl and rns) genes. The 15 conserved protein-coding genes include seven subunits of NADH dehydrogenase (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), thee subunits of cytochrome c oxidase (cox1, cox2, and cox3), three subunits of ATPase (atp6, atp8, and atp9), apocytochrome b (cob), and ribosomal protein S3 (rps3). The 24 tRNA genes cover all 20 standard amino acids and range in size from 71 to 84 bp.
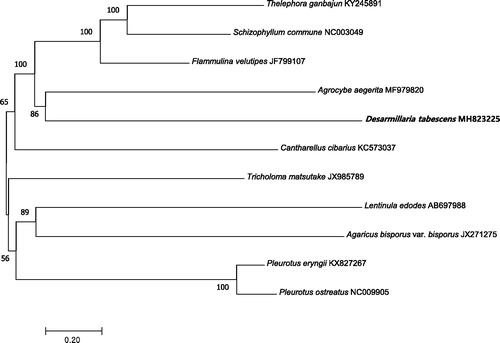
Phylogenetic analysis was conducted for the mitogenome and 10 other mushroom species using the neighbour-joining method of MEGA 7.0 with 1000 bootstrap replicates (Kumar et al. Citation2016). The phylogenetic tree indicated that the mitogenome of this species was genetically closest to that of Agrocybe aegerita (). The mitogenome of D. tabescens can contribute to our understanding of the phylogeny and evolution of this recently reassigned species.
Figure 1. Neighbour-joining phylogenetic tree based on the mitochondrial genome sequences of D. tabescens and other 10 mushroom species using MEGA 7.0 (Kumar et al. Citation2016).
Disclosure statement
The authors report no conflicts of interest. The sequence has been submitted to NCBI under the accession no. MH823225.
Additional information
Funding
References
- Amiri A, Bussey KE, Riley M, Schnabel G. 2008. Propiconazole inhibits Armillaria tabescens in vitro and translocates into peach roots following trunk infusion. Plant Dis. 92:1293–1298.
- Baumgartner K, Fujiyoshi P, Ledbetter C, Duncan R, Kluepfel D. 2018. Screening almond rootstocks for sources of resistance to Armillaria root disease. HortScience. 53:4–8.
- Coetzee MPA, Wingfield BD, Zhao J, van Coller SJ, Wingfield MJ. 2015. Phylogenetic relationships among biological species of Armillaria from China. Mycoscience. 56:530–541.
- Hasegawa E, Ota Y, Hattori T, Sahashi N, Kikuchi T. 2011. Ecology of Armillaria species on conifer in japan. Forest Pathol. 41:429–437.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655
- Koch RA, Wilson AW, Séné O, Henkel TW, Aime MC. 2017. Resolved phylogeny and biogeography of the root pathogen Armillaria and its gasteroid relative, Guyanagaster. BMC Evol Biol. 17:33.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33:1870–1874.
- Lu ZM, Tao WY, Zou XL, Fu HZ, Ao ZH. 2007. Protective effects of mycelia of Antrodia camphorata and Armillariella tabescens in submerged culture against ethanol-induced hepatic toxicity in rats. J Ethnopharmacol. 110:160–164.
- Park KH, Oh SY, Park MS, Kim MS, Klopfenstein NB, Kim NK, Park JY, Kim JJ, Han SK, Lee JK, Lim YW. 2017. Re-evaluation of Armillaria and Desarmillaria in South Korea based on ITS/tef1 sequences and morphological characteristics. Forest Pathol. 47:e12447.
- Sterry P, Hughes B. 2009. Collins complete British mushrooms and toadstools. London: HarperCollins.
- Terashita T, Chyuman S. 1987. Fungi inhabiting wild orchids in Japan (IV). Armillariella tabescens, a new symbiont of Galeola septentrionalis. Trans Mycol Soc Jpn. 28:145–154.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
