Abstract
The present study provides a mitochondrial genome sequence of Scytosiphon lomentaria, which is better than the previously established data in China. This mitogenome is 39,971 bp in length and consists of 35 protein-coding genes, 3 ribosomal-RNA genes, 24 transfer-RNA genes, and 5 open reading frames. Overall, the base composition was as follows: A, 29.7%; C, 12.8%; G, 19.2%; and T, 38.4%. The phylogenetic analysis based on the protein-coding genes indicated that the S. lomentaria collected in the samples was at an earlier differentiation position. These results contribute to the further understanding of Phaeophyceae phylogenetic relationships and species identification.
Scytosiphon lomentaria is a common brown seaweed, widely distributed in cold and warm waters around the world (Mann Citation1991; Kogame et al. Citation2005). This species belongs to the family Phaeophyceae of the order Ectocarpales, and grows attached to shells and stones in intertidal or shallow subtidal zones. Previous studies on its morphogenesis, life history, and molecular phylogenesis have suggested that the population of this species in different geographical areas vary substantially (Kogame, Rindi, et al. Citation2015). Earlier phylogenetic analyses based on the mitochondrial cox3 and the nuclear ribosomal ITS divided the North-East Atlantic and the Mediterranean populations into four separate well-supported clades (Kogame, Ishikawa, et al. Citation2015). Due to its delicious taste and rich nutritive value, S. lomentaria is a favorite food for the coastal residents of North-East Asia (Sahoo and Seckbach Citation2015). However, the quantity of S. lomentaria has been considerably reduced due to ecological damage and changes in the environmental factors. Establishing a complete mitochondrial genome sequence will contribute to the development of a strategy for the conservation of this important macroalgae.
In the present study, the complete mitochondrial genome of S. lomentaria was initially assembled from previously published Illumina sequencing data (SRR5026350), used as a PCR primer guide. These specimens were collected from the beach of Xinghai Bay, China. The whole mitochondrial genome was revised with 48 pairs of primers, which were amplified and sequenced by protocols in Gene Denovo Laboratory, where the specimen and DNA were stored. The length of the complete mitochondrial genome of S. lomentaria is 39,971 bp, with an A + T content of 68.1% and the following base composition: A (29.7%), C (12.8%), G (19.2%), and T (38.4%). The genome comprised 67 genes, including 35 protein-coding genes (PCGs), 24 tRNAs, 3 rRNAs, and 5 open reading frames (ORFs). The 24 tRNA genes were as follows: trnK(uuu), trnA(ugc), trnD(guc), trnM(cau), trnL(uaa), trnH(gug), trnC(gca), trnN(guu), trnF(gaa), trnW(cca), trnM(cau), trnQ(uug), trnL(uag), trnL(caa), trnG(gcc), trnY(gua), trnR(ucu), trnI(gau), trnE(uuc), trnQ(cug), trnS(gcu), tRNA-Ser, trnM(cau), trnP(ugg). The 35 protein-coding genes include (rps2-4, 7, 8, 10-14, and 19; rpl2, 5, 6, 14, 16, 31, and 1-7, 4L, 9, 11, cob, cox1-3, atp6, 8, 9, and tatC). Five ORFs contained ORF78, ORF40, ORF127, ORF227, and ORF757. Compared with the only other S. lomentaria mitogenome established in Sanggou Bay, China (Liu and Pang Citation2014), despite the identical number of annotation genes, there was an increase in the length of 3,053 bp, 1 ORF, and 1 tRNA less in the new S. lomentaria mitogenome sequenced by us.
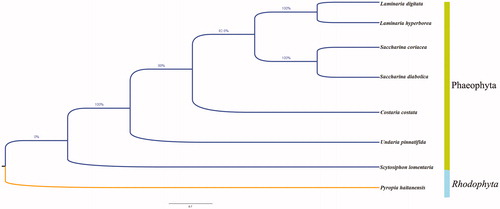
The phylogenetic relationship of S. lomentaria and other brown algae species was inferred using the neighbor-joining method in MEGA 7.0 (Kumar et al. Citation2016). The results showed that the species of Phaeophyceae were gathered in the same branch. Scytosiphon lomentaria was at an earlier differentiation position, and the distance between S. lomentaria and Undaria pinnatifida was nearer compared to those between other species of Phaeophyceae (). These new mitochondrial genome data are more complete and can be better used to provide a basis for studies of the mitochondrial evolution of Phaeophyceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kogame K, Ishikawa S, Yamauchi K, Uwai S, Kurihara A, Masuda M. 2015. Delimitation of cryptic species of the Scytosiphon lomentaria complex (Scytosiphonaceae, Phaeophyceae) in Japan, based on mitochondrial and nuclear molecular markers. Phycol Res. 63:167–177.
- Kogame K, Rindi F, Peters AF, Guiry MD, 2015. Genetic diversity and mitochondrial introgression in Scytosiphon lomentaria (Ectocarpales, Phaeophyceae) in the north-eastern Atlantic Ocean. Phycologia. 5:367–374.
- Kogame K, Uwai S, Shimada S, Masuda M. 2005. A study of sexual and asexual populations of Scytosiphon lomentaria (Scytosiphonaceae, Phaeophyceae) in Hokkaido, northern Japan, using molecular markers. European Journal of Phycology. 40:313–322.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Liu F, Pang S. 2014. Complete mitochondrial genome of the brown alga Scytosiphon lomentaria (Scytosiphonaceae, Phaeophyceae). Mitochondrial DNA. 27:1494–1496.
- Mann KH. 1991. Seaweeds: their environment, biogeography, and ecophysiology. by K. Luning. Limnol Oceanogr. 36:378–380.
- Sahoo D, Seckbach J. 2015. The algae world. Dordrecht: Springer Netherlands.