Abstract
Salinator takii is a marine gastropod mollusk belonging to the family Amphibolidae. In this study, we report the complete mitochondrial genome of S. takii. The 13,958 bp mitochondrial genome covers 13 protein-coding genes, 2 rRNAs, and 22 tRNAs, it is the shortest of pulmonate mitochondrial genome currently available in the Genebank. The GC% across the genome was 34.20%.
Salinator takii belong to the family Amphibolidae, which is distributed in most areas of Asia and the northern shore of the Indian ocean as well as Australia (Golding Citation2011). They live on the surface of soft, muddy substrate in estuarine salt marshes, mudflats, and mangrove forests, and are believed to be capable of both aerial and aquatic respiration during tidal immersion (Pilkington et al. Citation1984). For a long time, the study of higher relationships of pulmonate gastropods was difficult due to high morphological homoplasy and low sampling (White et al. Citation2011). Therefore, the complete mitochondrial genome of S. takii is necessary to shed some light on deeper levels of phylogenetic analyses of pulmonates.
The sample of S. takii was collected from Beihai, Guangxi Province, China (N 21°36′40.43″ E 108°44′20.23″), and preserved in 95% ethanol, at the Laboratory of Shellfish Genetics and Breeding, Ocean University of China. The total genomic DNA was extracted using cetyltrimethylammonium bromide (CTAB) extraction method (Schulenburg et al. Citation2001). Total genomic DNA was sequenced on an Illumina MiSeq sequencer using a PE150 protocol. Short-read DNA sequences were assembled using CLC Genomics Workbench 11 (CLC Bio, Aarhus, Denmark). The sequence data were annotated using MITOS (Bernt et al. Citation2013) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), gene boundaries were compared manually with published Amphibolidae mitochondrial genome.
The mitochondrial genome of S. takii is 13,958 bp long and was deposited in the Genebank (Accession number: MH588520). It is the shortest of 10 pulmonates mitochondrial genomes available in the Genebank. Thirteen protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes were found, consistent with other gastropod mitochondrial genomes. The rrnL and rrnS genes are 1012 and 699 bp in length, respectively. Overall nucleotide composition is as follows: A = 27.5%, C = 14.1%, G = 20.1%, and T = 31.8%. The GC skew value is 17.5%.
Some of the PCGS have adjacent overlapping region. The amount of overlap between genes is typically between 1 and 22 bp. Eight of the 13 PCGs start with TTG, while others start with GTG(2) and ATG(3). Five genes are terminated by TAG, two of which are terminated by TAA, but COX2, COX3, CYTB, ATP6, ATP8, and ND3 are terminated by incomplete stop codon T. Most of the PCGs were encoded on the (+) strand, while ATP8, ATP6, ND3, and COX3 were encoded on the (−) strand.
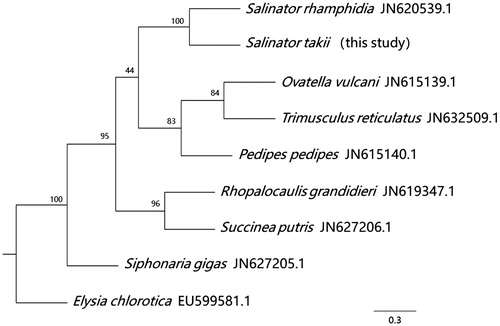
To further validate the new sequence, we used eight mitochondrial genomes from seven families of pulmonate to construct a maximum likelihood (ML) tree (), and Elysia chlorotica was used as the outgroup. Our sequence phylogenetically clustered with S. rhamphidia, contrary to the traditional view, suggesting that Amphibolinae is not basal with respect to all pulmonates.
Figure 1. Maximum likelihood tree was constructed based on the concatenated amino acid sequences of the 13 protein-coding genes. Maximum likelihood analysis was conducted using RAxML (Stamatakis Citation2014) and 1000 bootstraps were used to assess the support of nodes. The GTRGAMMA model was employed using RAxML.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Golding RE. 2011. Molecular phylogenetic analysis of mudflat snails (Gastropoda: Euthyneura: Amphiboloidea) supports an Australasian centre of origin. Mol Phylogenet Evol. 63:72–81.
- Pilkington JB, Little C, Stirling PE. 1984. A respiratory current in the mantle cavity of Amphibola crenata (Mollusca, Pulmonata). J Royal Soc New Zealand. 14:327–334.
- Schulenburg JHGV, Hancock JM, Pagnamenta A, Sloggett JJ, Majerus MEN, Hurst GDD. 2001. Extreme length and length variation in the first ribosomal internal transcribed spacer of ladybird beetles (Coleoptera: Coccinellidae). Mol Biol Evol. 18:648–660.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- White TR, Conrad MM, Tseng R, Balayan S, Golding R, Martins AMDF. 2011. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evol Biol. 11:295.
