Abstract
Heptacodium miconioides Rehd. is a rare and endangered plant species occurring in Anhui, Hubei, and Zhejiang Provinces of China, which is endemic to China. Here, we report and characterize the complete plastid genome sequence of H. miconioides in an effort to provide genomic resources useful for promoting its conservation. The complete plastome is 156,313 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat (IR) regions of 23,904 bp, a large single-copy (LSC) region of 89,760 bp and a small single-copy (SSC) region of 18,745 bp. The plastome contains 114 genes, comprising 79 unique protein-coding genes, 31 unique tRNA genes, and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA, and 16S rRNA). The overall A/T content in the plastome of D. indica is 61.60%. The complete plastome sequence of H. miconioides will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Caprifoliaceae.
Heptacodium miconioides Rehd. (Caprifoliaceae) is a shrub growing 7 m tall, which is endemic to China. It is widely distributed in Anhui, Hubei, and Zhejiang Provinces of China and it grows in cliffs, scrub, and forests with altitude from 600 to 1000 m (Yang et al. Citation2011). It is a rare and Endangered (EN) plant species (Qin et al. Citation2017). It can be used as an excellent ornamental species (Bian et al. Citation2002). In this study, H. miconioides was sampled from Hangzhou Botanical Garden in Zhejiang Province of China (120.112°E, 30.253°N). A voucher specimen (Wang et al., B158) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Kolkwitzia amabilis (KT966716.1) (Bai et al. Citation2017) using MITO bim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Lonicera japonica (KJ170923.1) (He and Qian Citation2014). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of H. miconioides is found to possess a total length 156,313 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 23,904 bp, a large single-copy (LSC) region of 89,760 bp, and a small single-copy (SSC) region of 18,745 bp. The plastome contains 114 genes, consisting of 79 unique protein-coding genes (three of which are duplicated in the IR: rps12, rps7, and ndhB), 31 unique tRNA genes (eight of which are duplicated in the IR), and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA and 16S rRNA). Among these genes, 8 pseudogenes (accD, clpP, rpoA, rps3, rpl23, ycf2, rps15, ycf1), 13 genes possessed a single intron, and three genes (ycf3, rps18, and rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of H. miconioides is 61.60%, for which the corresponding value of the LSC, SSC, and IR region were 63.40%, 67.10%, and 56.20%, respectively.
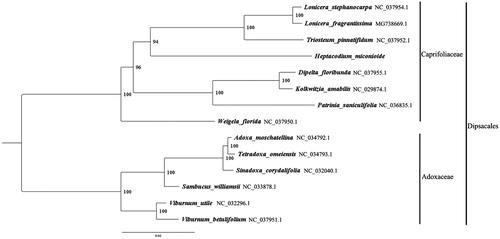
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of seven published complete plastomes of Caprifoliaceae: L. stephanocarpa, L. fragrantissima, Triosteum pinnatifidum, Dipelta floribunda, K. amabilis, Patrinia saniculifolia, Weigela florida, using six published complete plastomes of Adoxaceae as outgroups: Viburnum utile, V. betulifolium, Adoxa moschatellina, Tetradoxa omeiensis, Sinadoxa corydalifolia, Sambucus williamsii .The phylogenetic analysis indicated that H. miconioides is closer to Triosteum pinnatifidum than other taxa in this study (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of H. miconioides will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Caprifoliaceae.
Figure 1. The best ML phylogeny recovered from 14 complete plastome sequences by RAxML. Accession numbers: Heptacodium miconioides Rehd. (This study, Genbank accession number: MH712480), Lonicera stephanocarpa NC_037954.1, Lonicera fragrantissima MG_738669.1, Triosteum pinnatifidum NC_037952.1, Dipelta floribunda NC_037955.1, Kolkwitzia amabilis NC_029874.1, Patrinia saniculifolia NC_036835.1, Weigela florida NC_037950.1; outgroups:Adoxa moschatellina NC_034792.1, Tetradoxa omeiensis NC_034793.1, Sinadoxa corydalifolia NC_032040.1, Sambucus williamsii NC_033878.1, Viburnum utile NC_032296.1, Viburnum betulifolium NC_037951.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bai GQ, Zhou T, Zhao JX, Li W, Han GJ, Li SF. 2017. The complete chloroplast genome of Kolkwitzia amabilis (Caprifoliaceae), an endangered horticultural plant in China. DNA Sequence. 28:296–297.
- Bian CM, Jin ZX, Li JM. 2002. A study on the reproductive biology of Heptacodium miconioides. Acta Bot Yunnanica. 24:613–618.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads –– a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- He L, Qian J. 2014.The complete chloroplast genome sequence of Lonicera japonica. National Traditional Chinese Medicine and Natural Drugs Proseminar; August 26–29; Beijing, China.
- Qin H, Zhao L, Yu S, Liu H, Liu B, Xia N, Peng H, Li Z, Zhang Z, He X, et al. 2017. A list of threatened species of higher plants in China. Biodiversity. 25:697–744.
- Stamatakis A. 2006. RAxML-VI-HPC maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang QL, Landrein S, Osborne J, Borosova R. 2011. Flora of China. Vol. 19. Beijing: Science Press.
