Abstract
Pandanus tectorius Parkinson ex Du Roi (Pandanaceae) is a tree or shrub which is widely distributed in seashores and sandy beaches in Fujian, Guangdong, Guangxi, Guizhou, Hainan, Taiwan, Yunnan Provinces of China. P. tectorius sometimes is used as a living fence, the leaves are used for weaving and the fruit is beneficial for the treatment of diabetes. Here, we report and characterize the complete plastid genome sequence of P. tectorius in an effort to provide genomic resources useful for promoting its conservation. The complete plastome is 159,362 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat (IR) regions of 26,702 bp, a large single-copy (LSC) region of 87,447 bp and a small single-copy (SSC) region of 18,511 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA, and 16S rRNA). The overall A/T content in the plastome of P. tectorius is 62.40%. The complete plastome sequence of P.tectorius will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Pandanaceae.
Pandanus tectorius Parkinson ex Du Roi (Pandanaceae) is a tree or shrub which is widely distributed in seashores and sandy beaches in Fujian, Guangdong, Guangxi, Guizhou, Hainan, Taiwan, Yunnan provinces of China. P. tectorius sometimes is used as a living fence, and the leaves are used for weaving (Sun and Robert Citation2010). In addition, the P. tectorius fruit is beneficial for the treatment of diabetes (Wu et al. Citation2014). P. tectorius was sampled from the greenhouse within Hainan University campus, Haikou, Hainan, China (110.327°E, 20.059°N). A voucher specimen (Wang et al., B247) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Carludovica palmata (NC_026786.1) (Mennes et al. Citation2015) using MITO bim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Carludovica palmata (NC_026786.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of P. tectorius is found to possess a total length 159,362 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 26,702 bp, a large single-copy (LSC) region of 87,447 bp and a small single-copy (SSC) region of 18,511 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes (seven of which are duplicated in the IR: rps12, rps7, ndhB, ycf2, rpl23, rpl2 and rps19), 30 unique tRNA genes (eight of which are duplicated in the IR: trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA, trnI-CAU and trnH-GUG) and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA and 16S rRNA). Among these genes, three pseudogenes (rpoC2, ycf2 and ycf1), 15 genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB and ndhA) possessed a single intron and three genes (ycf3, clpP and rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of P. tectorius is 62.40%, which the corresponding value of the LSC, SSC and IR region were 64.40%, 68.10% and 57.20%, respectively.
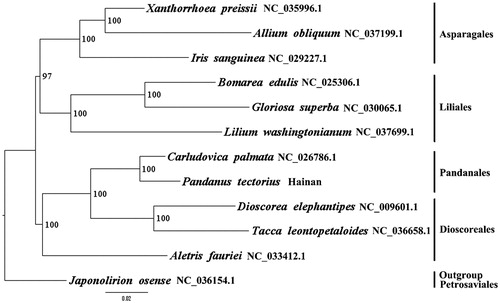
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of one published complete plastome of Pandanales, three published complete plastomes of Asparagales, three published complete plastomes of Liliales, three published complete plastomes of Dioscoreales, using Japonolirion osense (Petrosaviaceae, Petrosaviales) as outgroup. The phylogenetic analysis indicated that P. tectorius is closer to Carludovica palmata than other taxa in this study (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of P. tectorius will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Pandanaceae.
Figure 1. The best ML phylogeny recovered from 12 complete plastome sequences by RAxML. Accession numbers: Pandanus tectorius (this study, GenBank Accession number:MH748568), Carludovica palmata NC_026786.1, Xanthorrhoea preissii NC_035996.1, Allium obliquum NC_037199.1, Iris sanguinea NC_029227.1, Bomarea edulis NC_025306.1, Gloriosa superba NC_030065.1, Lilium washingtonianum NC_037699.1, Dioscorea elephantipes NC_009601.1, Tacca leontopetaloides NC_036658.1, Aletris fauriei NC_033412.1; outgroup: Japonolirion osense NC_036154.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Mennes CB, Lam VKY, Rudall PJ, Lyon SP, Graham SW, Smets EF, Merckx VSFT. 2015. Ancient Gondwana break-up explains the distribution of the mycoheterotrophic family Corsiaceae (Liliales). J Biogeogr. 42:1123–1136.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Sun K, Robert AD. 2010. Flora of China. Vol. 23, Beijing: Science Press.
- Wu CM, Zhang XP, Zhang X, Luan H, Sun GB, Sun XB, Wang XL, Guo P, Xu XD. 2014. The caffeoylquinic acid-rich Pandanus tectorius fruit extract increases insulin sensitivity and regulates hepatic glucose and lipid metabolism in diabetic db/db mice. J Nutr Biochem. 25:412–419.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
