Abstract
Pemphis acidula is a rare shrub in China and valued for its hard wood, bonsai production, and scientific research. Growing on beaches and in mangrove habitats, P. acidula populations are threatened by the rapid coastal reclamation and pollution around the world. In this study, we assembled and characterized the complete chloroplast genome of P. acidula as a resource for future genetic studies. Its chloroplast genome was 160,054 bp in size, with a large single-copy (LSC) region of 89,785 bp, a small single-copy (SSC) region of 18,883 bp, separated by two inverted repeat (IR) regions of 25,693 bp each. A total of 132 genes were predicted. Phylogenetic analysis showed a close relationship between P. acidula and Punica granatum, both are members of the family Lythraceae.
Pemphis acidula Forst is a tropical sea-shore plant species, distributed from East Africa to the Pacific Ocean. Its morphology can vary from a scrambler to a tall shrub and grows on raised coral and rocky or sandy beaches above the intertidal zone (Lewis and Rao Citation1971; Ellison et al. Citation2010). While traditionally harvested for its wood, the species is also valued as a bonsai species (Ellison et al. Citation2010) and is recently shown to be useful against mosquitoes (Samidurai et al. Citation2009). In addition to the bonsai trade, P. acidula populations are experiencing serious decline due to its affiliation with the mangrove environment that is rapidly being developed (Ellison et al. Citation2010). The species is the type species of Pemphis, and until recently was thought to be the only species in the genus (Lewis and Rao Citation1971). It is very important to do research on the phylogenetic relationship within Lythraceae and molecular mechanisms underlying plant salt or other stress tolerance in intertidal zone or coral islands. Herein, we characterized the complete chloroplast genome sequence of P. acidula as a resource for future genetic studies.
Fresh leaves of P. acidula were sampled from a live plant kept at the South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China. A voucher specimen was also deposited at the Herbarium of the South China Botanical Garden, Chinese Academy of Sciences (IBSC) with the accession number JianSG-201801. After DNA extraction, a library with insertion size of ∼500 bp was constructed, and high-throughput DNA sequencing (paired-end 150 bp) was performed on an Illumina Hiseq2500 platform. Approximately 4 Gb of sequence data were generated. Using a partial rbcL gene sequence of Lagerstroemia indica (GenBank accession NC_030484) as seed, the chloroplast genome was assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome sequence was then annotated using DOGMA (Wyman et al. Citation2004) and manually corrected.
The complete chloroplast genome sequence of P. acidula (GenBank accession MH727532) was 160,054 bp in length, with a large single-copy (LSC) region of 89,785 bp, a small single-copy (SSC) region of 18,883 bp, separated by a pair of inverted repeat (IR) regions of 25,693 bp each. There were 132 predicted genes, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was 36.46%.
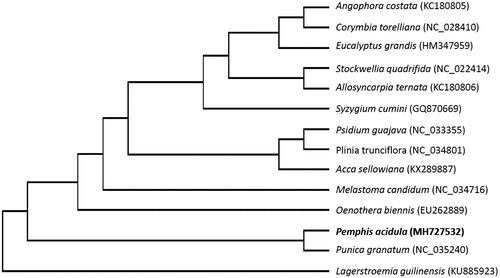
A phylogenetic analysis was conducted to understand the relationship of P. acidula within the order Myrtales. For that, 13 representative species of the order Myrtales were downloaded from the NCBI GenBank database, and aligned using MAFFT v7.307 (Katoh and Standley Citation2013). Using RAxML (Stamatakis Citation2014), a maximum likelihood tree was constructed with the GTR + G model, with Lagerstroemia guilinensis as outgroup. As shown in , P. acidula was found to be sister to Punica granatum (NC_035240), both members of the family Lythraceae, with strong bootstrap support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Ellison J, Koedam NE, Wang Y, Primavera J, Ong JE, Yong WHJ, Ngoc VN. 2010. Pemphis acidula. The IUCN Red List of Threatened Species. 2010:e.T178838A7622565.Cambridge, UK:Fauna & Flora International.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lewis D, Rao AN. 1971. Evolution of dimorphism and population polymorphism in Pemphis acidula Forst. Proc Royal Soc B. 178:1050.
- Samidurai K, Jebanesan A, Saravanakumar A, Govindarajan M, Pushpanathan T. 2009. Larvicidal, ovicidal and repellent activities of Pemphis acidula Forst. (Lythraceae) against filarial and dengue vector mosquitoes. Acad J Entomol. 2:62–66.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.