Abstract
The dwarf sperm whale is a small, toothed cetacean, which was suggested to be two separate species based on the evidence derived from partial mitochondrial DNA sequence. Here, the complete mitochondrial genome of Kogia sima (16,379 bp in length) has been analyzed for building the database. It was similar to the typical mammalian mtDNA, containing 37 genes (13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes) and a non-coding region. Overall base composition of the complete mitochondrial DNA is A (31.2%), G (14.4%), C (29.4%), T (25.0%), the percentage of A and T (56.2%) is higher than G and C (43.8%). All genes in K. sima were distributed on the H-strand, except for the ND6 subunit gene and 9 tRNA genes which were encoded on the L-strand. The phylogenetic relationships of 15 Odontoceti species were reconstructed based on the 13 protein-coding genes using the Bayesian inference method.
The dwarf sperm whale (Kogia sima Owen, Citation1866) is a small, toothed cetacean, which is distributed widely in offshore waters of tropical and warm temperate zones across the Atlantic and Indo-Pacific Ocean (Chivers et al. Citation2005; Mcalpin Citation2009). It shares the Kogia genus with only one extant species, the pygmy sperm whale (K. breviceps Blainville, 1838), which is larger in both total body length and weight than K. sima (Mcalpin Citation2009). Although there is genetic evidence to suggest that there may be two separate species of Dwarf Sperm Whales, one in the Atlantic and one in the Indo-Pacific Ocean (Chivers et al. Citation2005), the K. Sima is now treated as one species due to the lack of robust supporting evidence. To date, only partial sequence of the mtDNA of K. sima is available. Considering the small size of mitochondrial genome, maternal inheritance in most species, accelerated rate of mutation compared to the nuclear DNA and little or no recombination, it represents a mainstay of phylogenetic relationships resolving at several taxonomic levels (Brown et al. Citation1979; Ballard and Whitlock Citation2004). We determined the complete mitochondrial genome of K. sima for building the database which will be useful for resolving the taxonomy of dwarf sperm whales in the future, and analyzed the phylogenetic relationships between this species and other Odontoceti species.
The K. sima tissue specimen used in this study was collected from the carcass of a stranded individual in Xiamen coastal, Fujian Province (China), which is numbered 200623 and stored at the College of Life Sciences, Nanjing Normal University. The genomic DNA was extracted from the muscle tissue using TreliefTM Animal Genomic DNA Kit (TsingKe) according to the manufacturer’s instructions. The complete genome sequence of the K. sima (accession no. MH791441) is 16,379 bp in length which was sequenced by High-throughput sequencing technology (Illumina HiSeq platform). It encoded 37 genes totally containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding region (D-loop). All the genes in K. sima were distributed on the H-strand, except for the ND6 subunit gene and nine tRNA genes which were encoded on the L-strand. Overall base composition of the complete mitochondrial DNA is A (31.2%), G (14.4%), C (29.4%), and T (25.0%), which indicated a strong AT bias (Shadel & Clayton Citation1997).
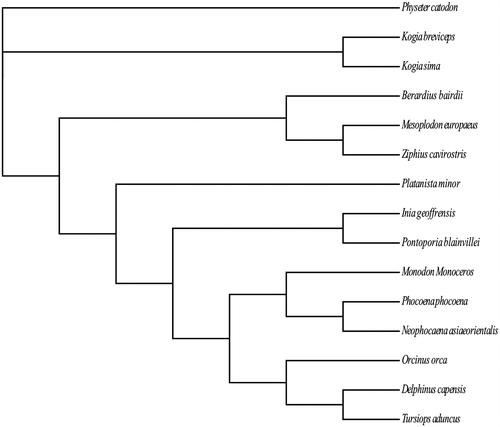
In order to confirm the mitochondrial sequence obtained in this study, we selected another 14 Odontoceti species to reconstruct the phylogenetic tree with the Bayesian inference method in the MrBayes version 3.2.2 (Huelsenbeck & Ronquist Citation2001). The topologies are completely consistent with the previous studies based on several mitochondrial and/or nuclear DNA markers, except for the position of Platanista and the sister relationship between the family Kogiidae and Physeteridae (McGowen et al. Citation2009; Steeman et al. Citation2009) (). The reason for these differences might be the accelerated rate of mutation on mtDNA compared to the nuclear DNA. In addition to these differences, others were completely in agreement with the tree of mitochondrial DNA sequences and the previous tree inferred from nuclear DNA. Hence, the result confirmed the phylogenetic relationships among Odontoceti, such as Kogiidae, Monodontidae, and Phocoenidae.
Figure 1. Phylogenetic relationships of 15 Odontoceti species based on concatenated amino acid sequences of the 13 PCGs. The tree is inferred from the Bayesian inference method with the general time reversible (GTR) model, and gamma distributed with invariant sites (G + I) as the substitution rate among sites. GenBank accession numbers for the published sequences are MH791441 (Kogia sima), NC_002503 (Physeter catodon), NC_005272 (Kogia breviceps), NC_005274 (Berardius bairdii), NC_005275 (Platanista minor), NC_005276 (Inia geoffrensis), NC_005277 (Pontoporia blainvillei), NC_005279 (Monodon monoceros), NC_005280 (Phocoena phocoena), NC_012058 (Tursiops aduncus), NC_012061 (Delphinus capensis), NC_021434 (Mesoplodon europaeus), NC_021435 (Ziphius cavirostris), NC_023889 (Orcinus orca), and NC_026456 (Neophocaena asiaeorientalis).
Acknowledgements
The authors are especially grateful to Dr. Yang and Dr. Chen for sample collection, and valuable suggestions on this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744.
- Brown WM, George M, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 76:1967–1971.
- Chivers S, Leduc RG, Robertson KM, Barros NB, Dizon AE. 2005. Genetic variation of Kogia spp. with preliminary evidence for two species of Kogia sima. Mar Mammal Sci. 21:619–634.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Mcalpin DF. 2009. Pygmy and dwarf sperm whales (Kogia breviceps and K. sima). In: Perrin WF, Würsig B, Thewissen JGM, editors. Encyclopedia of marine mammals. 2nd ed. San Diego, CA: Academic Press; p.936–938.
- McGowen MR, Spaulding M, Gatesy J. 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol Phylogenet Evol. 53:891–906.
- Owen R. 1866. On some Indian Cetacea collected by Walter Elliot. Esq. T Zool Soc Lond. 6:17–47.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Steeman ME, Hebsgaard MB, Fordyce RE, Ho SY, Rabosky DL, Nielsen R, Rahbek C, Glenner H, Sørensen MV, Willerslev E. 2009. Radiation of extant cetaceans driven by restructuring of the oceans. Syst Biol. 58:573–585.
