Abstract
Complete plastomes of 14 African yam species were reconstructed from whole genome sequencing. These plastomes sizes varied from 151,908 to 155,155 bp. We predicted 130 genes, including 18 duplicated genes located in the two inverted regions. Phylogenetic analysis obtained using the maximum likelihood procedure revealed that each species was distinct with high support. This resource will be useful for phylogenetic and diversity analysis of Dioscorea species.
Keywords:
The genus Dioscorea L. (Dioscoreaceae) plays a considerable role in tropical Africa because of its important staple crop D. rotundata, a vine cultivated for its starchy tuber. Dioscorea is a very species-rich and morphologically complex genus. Therefore, the taxonomy of this genus is difficult, as proven by the variation in the number of accepted species names (Caddick et al. Citation2002; Wilkin et al. Citation2005; Govaerts et al. Citation2017). Molecular phylogenetic analyses focusing on the genus are also poorly resolved (Terauchi et al. Citation1992; Ramser et al. Citation1997; Chaïr et al. Citation2005). Here we reconstructed complete plastomes of 14 West-African species.
Fourteen Dioscorea leaf samples, (D. abyssinica, D. baya, D. bulbifera, D. burkilliana, D. cayenensis, D. dumetorum, D. hirtiflora, D. praehensilis, D. preussii, D. quartiniana, D. sagittifolia, D. sansibarensis, D. schimperiana, and D. togoensis), were collected from WAG and G herbaria (). DNAs were extracted from dried leaves according to Scarcelli et al. (Citation2006) and whole genome sequencing was performed as described in Mariac et al. (Citation2014) on an Illumina MiSeq (Illumina, San Diego, CA). For each sample, the whole plastome was reconstructed using MITObim 1.7 (Hahn et al. Citation2013) as explained in Magwé-Tindo et al. (Citation2018) with D. rotundata plastome as a reference (GenBank NC_024170.1). Annotations were recovered from the alignment of the target plastome to NC_024170.1 using Geneious Pro 4.8.5 with Mauve 2.2.0 plugin (Darling et al. Citation2010), then manually checked for start and stop codons (protein-coding genes) and by blasting the tRNA and rRNA sequences to GenBank nucleotide database (nr/nt, Altschul et al. Citation1990). Annotated plastomes were deposited into the GenBank database ().
Table 1. Characterization of the samples used for sequencing.
Plastome lengths varied from 151,908 bp (D. burkilliana) to 155,155 bp (D. baya) and exhibited a typical quadripartite structure with a LSC (Large Single Copy) and a SSC (Short Single Copy) separated by two IR (Inverted Region). The mean GC content was 37% and varied from 31% within the SSC to 43% within the IR. The 14 species presented the same genetic structure as the reference (NC_024170.1): 130 genes corresponding to 84 protein-coding genes, 38 tRNA and 8 rRNA. Among these genes, 18 were duplicated in the IR: 6 protein-coding genes, 8 tRNA and 4 rRNA. The only exception was D. quartiniana which missed the matK gene due to a mutation of the second codon into a stop codon. All species lost the rps16 gene.
A phylogenetic study was conducted using these 14 complete plastomes, with D. rotundata (NC_024170.1) as reference and D. elephantipes (NC_009601.1) as outgroup species. The 16 plastomes were aligned using MAFFT 7 (Katoh et al. Citation2002) and the alignment was cleaned using Gblocks 0.91b (Talavera and Castresana Citation2007). The phylogenetic tree was constructed using RAxML 8.2.9 (Stamatakis Citation2014) with 500 bootstraps. The phylogenetic tree obtained was well-resolved and revealed that each species was clearly distinct with strong support (), except within the cultivated species complex.
Figure 1. Phylogenetic tree of 16 African Dioscorea species constructed using maximum likelihood procedure and whole plastome sequences. Dioscorea elephantipes was included as outgroup species. Bootstrap values are indicated for each node. Each species is followed by its GenBank accession ID.
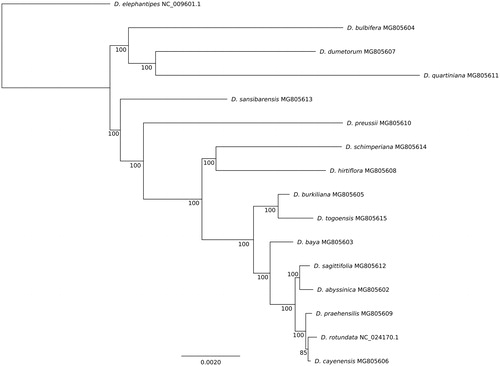
Resources reconstructed here will help to understand the relationships between cultivated yams and their wild relatives and provide useful information for further taxonomic study.
Acknowledgements
The authors thank the Conservatoire et Jardin botaniques de la ville de Genève (G) for the D. praehensilis sample.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Caddick LR, Rudall PJ, Wilkin P, Hedderson TAJ, Chase MW. 2002. Phylogenetics of Dioscoreales based on combined analyses of morphological and molecular data. Bot J Linn Soc. 138:123–144.
- Chaïr H, Perrier X, Agbangla C, Marchand JL, Daïnou O, Noyer JL. 2005. Use of cpSSRs for the characterisation of yam phylogeny in Benin. Genome. 48:674–684.
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 5:e11147.
- Govaerts R, Wilkin P, Raz L, Tellez-Valdes O. 2017. World checklist of Dioscoreales. Facilitated by the Royal Botanic Gardens, Kew. Published on the Internet; http://wcsp.science.kew.org (accessed 30 June 2017).
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Magwé-Tindo J, Wieringa JJ, Sonké B, Zapfack L, Vigouroux Y, Couvreur TLP, Scarcelli N. Forthcoming 2018. Guinea yam (Dioscorea spp.) wild relatives identified using whole plastome phylogenetic analyses. Taxon. 67(5):905–915.
- Mariac C, Scarcelli N, Pouzadou J, Barnaud A, Billot C, Faye A, Kougbeadjo A, Maillol V, Martin G, Sabot F, et al. 2014. Cost-effective enrichment hybridization capture of chloroplast genomes at deep multiplexing levels for population genetics and phylogeography studies. Mol Ecol Resour. 14:1103–1113.
- Ramser J, Weising K, Lopez-Peralta C, Terhalle W, Terauchi R, Kahl G. 1997. Molecular marker based taxonomy and phylogeny of Guinea yam (Dioscorea rotundata - D. cayenensis)). Genome. 40:903–915.
- Scarcelli N, Tostain S, Mariac C, Agbangla C, Da O, Berthaud J, Pham J-L. 2006. Genetic nature of yams (Dioscorea sp.) domesticated by farmers in Benin (West Africa). Genet Resour Crop Evol. 53:121–130.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577.
- Terauchi R, Chikaleke VA, Thottappilly G, Hahn SK. 1992. Origin and phylogeny of Guinea yams as revealed by RFLP analysis of chloroplast DNA and nuclear ribosomal DNA. Theor Appl Genet. 83:743–751.
- Wilkin P, Schols P, Chase MW, Chayamarit K, Furness CA, Huysmans S, Rakotonasolo F, Smets E, Thapyai C. 2005. A plastid gene phylogeny of the yam genus, Dioscorea: roots, fruits and Madagascar. Syst Botany. 30:736–749.
