Abstract
In this study, the complete 15,662 bp mitochondrial genome was determined from Uca borealis using next-generation sequencing technology. It consists of 13 protein-coding genes, 22 tRNAs, 2 ribosomal RNAs, and a control region. Among the 37 genes, 23 genes (9 PCGs and 14 tRNA genes) are encoded on the heavy (+) strand while the remaining 14 genes (four PCGs, eight tRNA genes and two rRNA genes) are located on the light (−) strand. The AT-skew (0.01) for the whole mitogenome is slightly positive while GC-skew (−0.25) is negative, indicating a higher occurrence of As than Ts and Cs than Gs. The phylogenetic tree was constructed using MrBayes method based on the PCGs showed the present species clustered within the Brachyura. The complete mitogenome data would be useful for further study of U. borealis.
Uca borealis (Crane 1975), belongs to the fiddler crab, is widely distributed on sandy shores (Shin et al. Citation2004). Fiddler crab (Brachyura, Ocypodidae, Uca) is a famous group on the beach, and this genus encompasses over 90 species and subspecies (Rosenberg Citation2001). Fiddler crabs are characterized by extreme form of sexual dimorphism and body symmetry (Rosenberg Citation2002). Fiddler crabs have been studied extensively, such as morphology, sexual selection, and foraging (Shih et al. Citation1999). However, to date, the genetics background about U. borealis was rarely reported. Here, we newly determined the complete mitochondrial genome of U. borealis.
The sequenced specimen was collected in Beihai (China) on June 25th, 2018 (21°29′17″N, 109°07′29″E). All samples were stored at the Jiangsu Provincial Key Laboratory of Coastal Wetland Bioresources and Environmental Protection, Yancheng Teachers University, Yancheng, Jiangsu Province, China. Total DNA was extracted from the muscle tissues and using the Aidlab Genomic DNA Extraction Kit (Aidlab Biotech, Beijing, China). The mitogenomes of U. borealis were sequenced by next-generation sequencing (Illumina HisSeq 4000; Shanghai Origingene Bio-pharm Technology Co. Ltd. China), and clean data without sequencing adapters were de novo assembled by the NOVOPlasty software (Dierckxsens et al. Citation2017). The complete mitogenome sequence of Uca borealis has a closed circular molecule 15,662 bp in size. The gene content is typical of other decapoda mitochondrial genomes, including 13 PCGs, 2 rRNA genes, 22 tRNA genes, and a major non-coding region known as the CR with length of 745 bp. Among the 37 genes, 23 genes (9 PCGs and 14 tRNA genes) are encoded on the heavy (+) strand while the remaining 14 genes (4 PCGs, 8 tRNA genes and 2 rRNA genes) are located on the light (−) strand. The base composition bias of the U. borealis mitogenome is 69.47% A + T (A = 35.1%, G = 11.5%, T = 34.4% and C = 19.0%), made up of 67.33% in the PCGs, 73.57% in the transfer RNA genes, 74.64% in the ribosomal RNA genes and 75.70% in the CR. The AT-skew (0.01) for the whole mitogenome is slightly positive while GC-skew (−0.25) is negative, indicating a higher occurrence of As than Ts and Cs than Gs. The mitogenome of U. borealis has been deposited in GenBank under accession number MH796170.
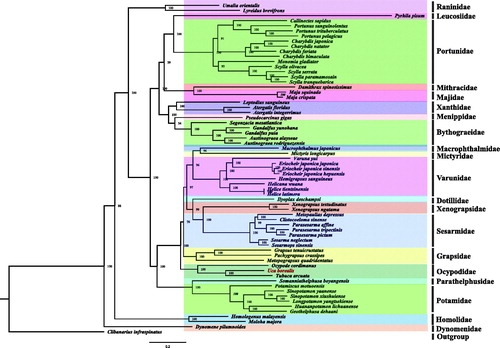
To reconstruct the phylogenetic relationship among crabs, the complete mitogenomes of other 62 Brachyura and one outgroup (Clibanarius infraspinatus) were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). The concatenated set of nucleotide sequences were used for phylogenetic analysis, which was performed with the BI method using MrBayes v3.2.6 (Huelsenbeck and Ronquist Citation2001). From the phylogenetic tree, we found U. borealis and Tubuca arcuata are clustered in one branch with high nodal support value (BI posterior probabilities [PP] = 100, ). In addition, the relationship among Sesarmidae, Xenograpsidae, Grapsidae, Ocypodidae, Varunidae, Potamidae, and Bythograeidae is also identified and is largely consistent with previous researches (Shen et al. Citation2013; Wang et al. Citation2018a, Citation2018b).
Figure 1. Phylogeny of Brachyura based on nucleotide sequences. The phylogenetic tree was inferred from the nucleotide sequences of 13 mitogenome PCGs using BI and ML methods. Numbers on branches indicate posterior probability (BI). Clibanarius infraspinatus was used as outgroup. The genbank accession numbers for all of the sequences is listed as follows: Atergatis floridus NC_037201.1, Atergatis integerrimus NC_037172.1, Austinograea alayseae NC_020314.1, Austinograea rodriguezensis NC_020312.1, Callinectes sapidus NC_006281.1, Charybdis feriata NC_024632.1, Charybdis japonica NC_013246.1, Charybdis natator NC_036132.1, Clibanarius infraspinatus NC_025776.1, Clistocoeloma sinense NC_033866.1, Dynomene pilumnoides KT182070.1, Eriocheir japonica hepuensis NC_011598.1, Eriocheir japonica japonica NC_011597.1, Eriocheir japonica sinensis NC_006992.1, Gandalfus puia NC_027414.1, Gandalfus yunohana NC_013713.1, Geothelphusa dehaani NC_007379.1, Grapsus tenuicrustatus NC_029724.1, Harybdis bimaculata MG489891.1, Helicana wuana NC_034995.1, Helice latimera NC_033865.1, Helice tientsinensis NC_030197.1, Hemigrapsus sanguineus NC_035307.1, Homologenus malayensis NC_026080.1, Huananpotamon lichuanense NC_031406.1, Ilyoplax deschampsi NC_020040.1, Leptodius sanguineus NC_029726.1, Longpotamon yangtsekiense NC_036946.1, Lyreidus brevifrons NC_026721.1, Macrophthalmus japonicus NC_030048.1, Maguimithrax spinosissimus NC_025518.1, Maja crispata NC_035424.1, Maja squinado NC_035425.1, Metopaulias depressus NC_030535.1, Metopograpsus quadridentatus MH310445, Mictyris longicarpus NC_025325.1, Moloha majora NC_029361.1, Monomia gladiator NC_037173.1, Ocypode cordimanus NC_029725.1, Ortunus pelagicus KR153996.1, Otamiscus motuoensis KY285013.1, Pachygrapsus crassipes NC_021754.1, Parasesarma affine MH310444, Parasesarma pictum MG580780, Parasesarma tripectinis NC_030046.2, Portunus pelagicus NC_026209.1, Portunus sanguinolentus NC_028225.1, Portunus trituberculatus NC_005037.1, Pseudocarcinus gigas NC_006891.1, Pyrhila pisum NC_030047.1, Scylla olivacea NC_012569.1, Scylla paramamosain NC_012572.1, Scylla serrata NC_012565.1, Scylla tranquebarica NC_012567.1, Segonzacia mesatlantica NC_035300.1, Sesarma neglectum NC_031851.1, Sesarmops sinensis NC_030196.1, Sinopotamon xiushuiense NC_029226.1, Sinopotamon yaanense NC_036947.1, Somanniathelphusa boyangensis NC_032044.1, Tubuca arcuata KX911977.1, Umalia orientalis NC_026688.1, Varuna yui NC_037155.1, Xenograpsus ngatama NC_035951.1, Xenograpsus testudinatus NC_013480.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Rosenberg MS. 2001. the systematics and taxonomy of Fiddler crabs: a phylogeny of the genus Uca. J Crust Biol. 21:839–869.
- Rosenberg MS. 2002. Fiddler crab claw shape variation: a geometric morphometric analysis across the genus Uca (Crustacea: Brachyura: Ocypodidae). Biol J Linn Soci. 75:147–162.
- Shen H, Braband A, Scholtz G. 2013. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mole Phyl Evol. 66:776–789.
- Shih HT, Mok HK, Chang HW, Lee SC. 1999. Morphology of Uca formosensis Rathbun, 1921 (Crustacea: Decapoda: Ocypodidae), an endemic fiddler crab from Taiwan, with notes on its ecology. Zoo Stud. 38:164–177.
- Shin PKS, Yiu MW, Cheung SG. 2004. Behavioural adaptations of the Fiddler crabs Uca vocans borealis (crane) and Uca lactea lactea (De Haan) for coexistence on an intertidal shore. Mar Fre Beha Phys. 37:147–160.
- Wang Z, Wang Z, Shi X, Wu Q, Tao Y, Guo H, Ji C, Bai Y. 2018a. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int J Biol Macromol. 118:31–40.
- Wang Z, Shi X, Tao Y, Wu Q, Bai Y, Guo H, Tang D. 2018b. The complete mitochondrial genome of Parasesarma pictum (Brachyura: Grapsoidea: Sesarmidae) and comparison with other Brachyuran crabs. Genomics. 1–24. doi: 10.1016/j.ygeno.2018.05.002.
