Abstract
Alismatales play an important role in the systematics of angiosperm, yet different phylogenetic topologies of Alismatales based on different species have been generated from several studies. Here we reported and characterized the first complete chloroplast (cp) genomes of two Alismataceae species, Alisma plantago-aquatica and Sagittaria trifolia, and analyzed the phylogenomic relationship of Alismatales based on the complete chloroplast sequences of 21 Alismatales species. The two chloroplast genomes have the typical quadripartite structure with 113 genes in total, including 79 protein-encoding genes, 30 transfer RNA genes and four ribosomal RNA genes. Both IR regeions of plastid genomes contain 8 protein-encoding genes, 7 tRNA genes and all the four rRNA genes. The genome of A. plantago-aquatica is 177,417 bp in length, with an overall G + C content of 35.7%, and the genome of Sagittaria trifolia is 167,642 bp in length, with an overall G + C content of 37.0%. Phylogenetic study showed that the three Alismataceae species comprise a monophyletic group with a high support rate, which was a sister to the Hydrocharitaceae clade. The core Alismatids, consisting of Potamogetonaceae, Zosteraceae, Hydrocharitaceae and Alismataceae, is sister to Tofieldiaceae. Alocasia macrorrhizos of Araceae is the basal taxon of Alismatales, and the other twelve Araceae species formed a sister group to the clade Tofieldaceae and core Alismatids.
Alismatales are regarded as one of the oldest lineages within monocots that play an important role in the systematics of angiosperm (Soltis Citation2005). In recent years, some studies have focused on the phylogenomics of Alimatales, yet, different phylogenetic topologies have been reported. Either Araceae or Tofieldiaceae was thought to be the basal group of Alismatales (Ross Citation2016; Luo 2016). To better understand the evolution of Alimatales, we reported and characterized the first complete cp genomes of two Alismataceae species, Alisma plantago-aquatica and Sagittaria trifolia, and then analyzed the phylogenomic relationship of Alismatales based on the complete chloroplast sequences of 21 Alismatales species.
Leaf samples of Alisma plantago-aquatica and Sagittaria trifolia were collected on the side of Irtysh River, Beitun city, Xinjiang, China. The corresponding voucher herbarium specimens (Yang2017003 & Yang2017003) were deposited at the Herbarium of Tarim University (TAU). Total DNA was extracted from the silica-gel dried leaf tissue using DNA Plantzol Reagent (Invitrogen, U.S.), following the manufacturer’s protocol. Then, next-generation sequencing was conducted on the Illumina Hiseq Platform (Illumina, San Diego, CA). The complete cp genomes were assembled using NOVOPlasty (Dierckxsens et al. Citation2017) with the cp genome sequence of Sagittaria lichuanensis (GenBank accession number: NC_029815) as the reference. Sequence annotation was added using Geneious 11.0.4 (Biomatters Ltd., Auckland, New Zealand). Finally, clean reads were re-mapped to the draft genomes, yielding the whole cp genome sequences of A. plantago-aquatica (GenBank accession number: MK090659) and S. trifolia (GenBank accession number: MK090658). The two chloroplast genomes have a typical quadripartite structure, which is consistent with plastid genomes of other Alismatales and most angiosperm species (Yang 2016). The plastid genomes of the two species contain the same 113 genes, including 79 encoding genes, 30 transfer RNA genes and four ribosomal RNA genes. The IR regeions harbor eight protein-encoding genes (rp12, rp123, ycf2, ndhB, rps7, ycf1, rps15, ndhH), 7 tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU) and all the four rRNA genes (rrn16, rrn23, rrn4.5 and rrn5). The ndhH gene was pseudogenized probably due to incomplete duplication of the IR regions. However, the genome length and G + C content are obviously different between the two species. The cp genome of A. plantago-aquatica is 177,417 bp in length, with an overall G + C content of 35.7%, while the cp genome of Sagittaria trifolia is 167,642 bp in length, with an overall G + C content of 37.0%.
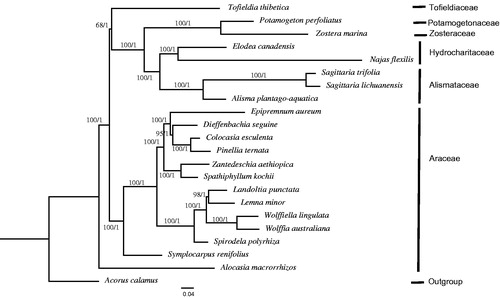
The phylogeny of Alismatales was reconstructed based on the complete cp genome of 21 Alismatales species and one outgroup taxon (Acorus calamus, Acoraceae) using the maximum likelihood (ML) and Bayesian inference (BI) methods implemented on the CIPRES Science Gateway V.3.3 (Miller et al. Citation2010). RAxML-HPC v.8.2.10 (Stamatakis Citation2014) and XSEDE v3.2.6 (Ronquist and Huelsenbeck Citation2003) are used for constructing ML and Mrbayes trees, respectively. As a result, the two analyses generated the same tree topology (). Three Alismataceae species, including two new-assembled chloroplast genomes in this study, comprise a group with a high support rate, sister to the Hydrocharitaceae clade. The core Alismatids, consisting of Potamogetonaceae, Zosteraceae, Hydrocharitaceae and Alismataceae, is sister to Tofieldiaceae. Alocasia macrorrhizos of Araceae is the basal taxon of Alismatales, and the other twelve Araceae species formed a sister group to the clade Tofieldaceae and core Alismatids. The results were largely consistent with a complete genus-level phylogeny of 13 Alismatales species based on plastid sequences (Nauheimer Citation2012), although the bootstrap value of the Alismatids and Tofieldiaceae clade was only 68%. On the other hand, Luo (Citation2016) reported that Tofieldiaceae is the basal group of Alismatales using plastid phylogenomic analyses based on 79 plastid protein-coding genes of ten Alismatales species. Further studies are required to resolve the deep phylogenetic relationship under Alismatales.
Figure 1. Molecular phylogeny of Alismatales based on the complete chloroplast genomes of 21 taxa, with Acorus calamus (Acoraceae) as the outgroup. The accession numbers are listed as below: Acorus calamus (AJ879453), Alocasia macrorrhizos (KR296655), Colocasia esculenta (JN105690), Dieffenbachia seguine (NC_027272), Elodea canadensis (NC_018541), Epipremnum aureum (KR872391), Lemna minor (DQ400350), Landoltia punctata (KY993962), Najas flexilis (NC_021936), Potamogeton perfoliatus (NC_029814), Pinellia ternata (NC_027681), Sagittaria lichuanensis (NC_029815), Spathiphyllum kochii (NC_030371), Spirodela polyrhiza (NC_015891), Symplocarpus renifolius (NC_033970), Tofieldia thibetica (KT899950), Wolffiella lingulata (JN160604), Wolffia australiana (JN160605), Zantedeschia aethiopica (NC_035499), Zostera marina (NC_036014). Relative branch lengths are indicated. Numbers above the lines represent ML bootstrap values/BI posterior probability. The hyphen indicates that a BI posterior probability or ML bootstrap <50%.
Disclosure statement
The authors are really grateful to the opened raw genome data from public GenBank. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Luo Y, Ma PF, Li HT, Yang JB, Wang H, Li DZ. 2016. Plastid phylogenomic analyses resolve Tofieldiaceae as the root of the early diverging monocot order Alismatales. Genome Biol Evol. 8:932–945.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans (LA):p. 1–8.
- Nauheimer L, Metzler D, Renner SS. 2012. Global history of the ancient monocot family araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytologist. 195:938–950.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Ross TG, Barrett CF, Gomez SM, Lam VKY, Henriquez CL, Les DH, Davis JI, Cuenca A, Petersen G, Seberg O, et al. 2016. Plastid phylogenomics and molecular evolution of alismatales. Cladistics. 32:160–178.
- Soltis DE, Soltis PS, Endress PK, Chase MW. 2005. Phylogeny and evolution of angiosperms. Sunderland, USA: Sinauer Associates.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
