Abstract
The complete mitochondrial genome sequence of Conanalus pieli was obtained by next-generation sequencing, and its phylogenetic status was analyzed. The complete mitogenome of C. pieli is 15,819 bp in length and typically consists of 37 genes, including 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one D-loop. All 13 protein-coding genes (PCGs) begins with ATN. Most of the PCGs used TAA as their stop codons, while the others used TAG or T as stop codons. The phylogenetic tree was constructed using PCGs and rRNAs of C. pieli and species from Tettigoniidae. The tree showed that C. pieli was grouped together with the three species that belong to the genus Conocephalus and the phylogenetic tree also supported the monophyly of Conocephalinae.
Keywords:
Conanalus pieli mainly lives in shallow mountainous areas that are approximately 1000 m above sea level, and feeds on other insects and small animals in bamboo forests (Xi and Zheng Citation1994). C. pieli belongs to Conocephalinae, Tettigoniidae, Orthoptera, and it is the type species of the genus Conanalus (Shi et al. Citation2005). The C. pieli samples were collected from Shimen, Hunan, China (N 29°58′, E 111°38′), on 18 August 2004, and was numbered as MSL0221 in the Molecular and Evolutionary Lab at Shaanxi Normal University. Total genomic DNA was extracted from the leg muscle of the samples. The genome of C. pieli was sequenced using next-generation sequencing technology. A total of 20,695,350 raw reads were generated from HiSeq (Illumina, San Diego, CA), and the length of one read was approximately 140 bp. Of these reads, 33,483 high quality reads were used to perform assembly using Mira 4.0.2 and MITObim 1.7 to produce a single, circular form of complete mitogenome (Hahn et al. Citation2013).
The complete mitogenome of C. pieli is 15,819 bp in length (GenBank accession number MH712457) and typically consists of 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one D-loop. The overall base composition of C. pieli mitogenome are A (38.1%), C (15.6%), G (10.2%) and T (36.1%), with an AT bias of 74.2%. PCGs were annotated by Geneious 10.1.2 (Biomatters Ltd., Auckland, New Zealand), and all 13 PCGs begin with ATN. COX2, COX3, ND5 and ND4 end with a single stop nucleotide T, CYTB with TAG, and other eight genes with TAA. rRNA and D-loop were also annotated using Geneious 10.1.2. 16S rRNA and 12S rRNA genes are 1,324 bp and 786 bp in length respectively, and D-loop was 966 bp, with a more obvious AT bias of 81.4%. The 22 tRNA genes, which were predicted by online software MITOS, range from 63 to 70 bp (Bernt et al. Citation2013).
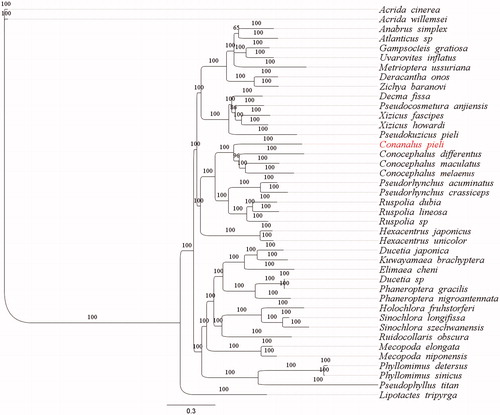
The phylogenetic tree was constructed based on a Bayesian inference method using 13 PCGs and two rRNA genes of 39 species from Tettigoniidae, including C. pieli in this study (). The phylogenetic analysis found that C. pieli was grouped together with three species from Conocephalus, which indirectly verifies the accuracy of C. pieli mitogenome sequencing, and the monophyly of Conocephalinae was supported by the phylogenetic tree ().
Figure 1. Phylogenetic tree generated based on 13 PCGs and two rRNAs using mitochondrial genomes from species in Tettigoniidae. The GenBank accession numbers for tree construction is listed as follows: Acrida cinerea (GU344100), Acrida willemsei (EU938372), Altbrus simplex (NC_009967), Atlanticus sp. (KX057730), Gampsocleis gratiosa (NC_011200), Uvarovites inflatus (NC_026231), Metrioptera ussuriana (NC_034796), Deracantha onos (NC_011813), Zichya baranovi (NC_033984), Decma fissa (NC_033981), Pseudocosmetura anjiensis (NC_033853), Xizicus fascipes (NC_018765), Xizicus howardi (KY458226), Pseudokuzicus pieli (NC_033982), Conocephalus differentus (MF347703), Conocephalus maculatus (HQ711931), Conocephalus melaenus (KY407794), Pseudorhynchus acuminatus (NC_033992), Pseudorhynchus crassiceps (NC_033990), Ruspolia dubia (EF583824), Ruspolia lineosa (NC_033991), Ruspolia sp. (KX057717), Hexacentrus japonicus (NC_033983), Hexacentrus unicolor (NC_033999), Ducetia japonica (KY612457), Kuwayamaea brachyptera (NC_028159), Elimaea cheni (NC_014289), Ducetia sp. (KX673198), Phaneroptera gracilis (NC_034756), Phaneroptera nigroantennata (NC_034757), Holochlora fruhstorferi (NC_033993), Sinochlora longifissa (NC_021424), Sinochlora szechwanensis (KX354724), Ruidocollaris obscura (NC_028160), Mecopoda elongata (NC_021380), Mecopoda niponensis (NC_021379), Phyllomimus detersus (NC_028158), Phyllomimus sinicus (NC_033997), Pseudophyllus titan (NC_034773) and Lipotactes tripyrga (NC_033996).
Acknowledgements
I thank Yuan Huang, Fuming Shi, and Shaoli Mao for the supply and identification of the specimen.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Shi FM, Wang JF, Peng F. 2005. A review of the genus Conanalus Tinkham (Orthoptera, Tettigonioidea) from China. Acta Zoota Sin. 30:84–86.
- Xi GS, Zheng ZM. 1994. Biological characteristics of Conanalus pieli (Orthoptera: Tettigonidae). J Shaanxi Norm Univ (Nat Sci Ed). 22:53–55.
