Abstract
The full-length mitochondrial genome of Pseudohelice subquadrata (Dana, 1851) was analyzed by the primer walking method. Its mitogenome is 16,884 bp in length, comprising 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. The phylogenetic tree confirmed that P. subquadrata belonged to the subsection Thoractotremata within the Crustacea. This is the first record of the complete mitogenome for the genus Pseudohelice.
Pseudohelice subquadrata (Dana, 1851) (Crustacea: Decapoda: Varunidae) is widely distributed in the tropic zones throughout the Indian Ocean to South Pacific Ocean (Shih & Suzuki Citation2008; Naderloo Citation2017; GBIF 2018). This varunid species is dominant at salt marshes and estuarine environments, especially intertidal zone and substrate mangroves (Sakai et al. Citation2006; Naderloo Citation2017). The Korea peninsula is the northern limit line of its natural distribution, and it was reported to occur at very restricted areas of southern coasts and Jeju Island of Korea (Kim Citation1973). Sakai et al. (Citation2006) separated the genus Pseudohelice Sakai et al. (Citation2006) from the Helice complex, based on morphological characters of endophragmal system and stridulating apparatus. Recent phylogenetic study based on partial mitochondrial sequences showed that the sisterhood relationship of P. subquadrata to Helice species (Sun et al. Citation2009).
A specimen of P. subquadrata was collected in the intertidal zone from Jeju Island, Korea, on 30 May 2017, after permission from the Ministry of Environment of Korea. The voucher specimen (MABIK GR00001388) was deposited in the collection of the National Marine Biodiversity Institute of Korea (MABIK) (Seocheon, Republic of Korea). Genomic DNA was extracted from its 5th walking leg tissue. To amplify the complete mitochondrial genome sequence, two independent and overlapping PCR runs were conducted with conserved primer sets newly designed in this study, and the two PCR amplicons were directly sequenced using the primer walking method. The full-length mitochondrial genome sequence was deposited in the GenBank under the accession number MH718959.
All mitochondrial genome sequences of the subsection Thoracotremata were retrieved from NCBI. They were aligned together with the P. subquadrata sequence analyzed in this study and refined manually. The nucleotide matrix of 13 protein-coding genes was created with the first, second and third positions of codon triplets. The final matrix consisted of 3705 bp for each codon position, respectively. Phylogenetic analysis was conducted using RAxML 7.0.4 (Stamatakis Citation2006) for maximum-likelihood (ML) analysis and MrBayes 3.1.2 (Ronquist et al. Citation2005) for Bayesian inference of phylogeny (BI). The alignment information is available upon request in FASTA format.
The complete mitochondrial genome sequence of P. subquadrata was a circular molecule of 16,884 bp in length, consisting of 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Its gene order was identical to the previously reported mitochondrial genomes of closely related species belonging to genera Helice and Helicana. This is the first report of a full-length mitochondrial genome sequence for the genus Pseudohelice.
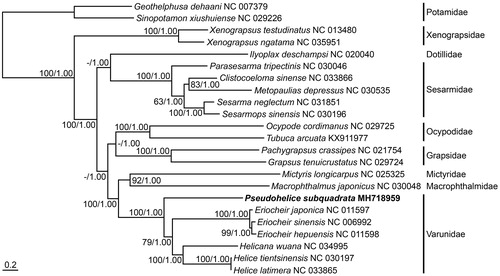
On the basis of the complete mitochondrial genome sequence of P. subquadrata analyzed in this study, a phylogenetic tree was reconstructed by the ML and BI methods, with the nucleotide sequence matrix from 13 protein-coding genes (). The reconstructed tree strongly supported the monophyly of all species belonging to the subsection Thoracotremata, with 100% bootstrap value in ML analysis and 1.00 posterior probability in BI analysis against the outgroups (i.e. two species belonging to the family Potamidae). Overall topology among the major families of the phylogenetic tree was mostly congruent with that of Basso et al. (Citation2017). Among them, all species belonging to the family Varunidae consistently formed a monophyletic group with high statistical supports. Within the clade, P. subquadrata placed at the most basal position and the other varunid species were further subdivided into three clades in accordance to their generic taxonomy, that is, Eriocheir, Helicana, and Helice. The latter two genera clustered more closely together and clearly separated from the former genus. The phylogenetic relationship of P. subquadrata based on its complete mitochondrial genome sequence was clearly different from that of Sun et al. (Citation2009) based on partial mitochondrial sequences and supported the taxonomic distinction of the genus Pseudohelice of Sakai et al. (Citation2006).
Figure 1. The phylogenetic tree of maximum-likelihood (ML) based on 13 protein-coding genes in the complete mitochondrial genome sequences from the species belonging to the subsection Thoracotremata. The matrix included the first, second, and third codon positions of their protein-coding genes. Two species belonging to the family Potamidae were used as outgroups. A bootstrap value above 50% in the ML analysis and posterior probability above 0.90 in the Bayesian analysis are indicated at each node. The varunid Pseudohelice subquadrata investigated in this study is shown in bold.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Basso A, Babbucci M, Pauletto M, Riginella E, Patarnello T, Negrisolo E. 2017. The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci Rep. 7:4096.
- Global Biodiversity Information Facility (GBIF). 2018. http://www.gbif.org/species/5863336.
- Kim HS. 1973. Illustrated encyclopedia of fauna and flora of Korea. Vol. 14. Anomura: Brachyura. Seoul: Samwha Publishing Co.; p. 1–649.
- Naderloo R. 2017. Atlas of crabs of the Persian Gulf. Cham: Springer. Family Varunidae H. Milne Edwards, 1853; p. 357–363.
- Ronquist F, Huelsenbeck JP, Van der Mark P. 2005. MrBayes 3.1.2: User manual. MrBayes 3.1, Florida.
- Sakai K, Türkay M, Yang SL. 2006. Revision of the Helice/Chasmagnathus complex (Crustacea: Decapoda: Brachyura). Abh Senckenberg Naturf Ges. 565:1–76.
- Shih H-T, Suzuki H. 2008. Taxonomy, phylogeny, and biogeography of the endemic mudflat crab Helice/Chasmagnathus complex (Crustacea: Brachyura: Varunidae) from East Asia. Zool Stud. 47:114–125.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Sun H, Jin Y, Zhang D, Yang S, Li Q, Song D, Zhou K. 2009. Mitochondrial sequence data reveals the phylogeny of the Asian Helice group of crabs (Decapoda: Brachyura: Varunidae). J Zool Syst Evol Res. 47:322–327.
