Abstract
The complete mitochondrial genome (mitogenome) sequence of the MeCord's box turtle (Cuora mccordi) was firstly determined by shotgun sequencing. The total length of mitogenome is 16,688 bp, and contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes and 2 main noncoding regions (the control region and the putative L-strand replication origin). The majority genes (12 protein-coding genes, 2 rRNA and 14 tRNA) were distributed on the H-strand, except for the ND6 subunit gene and eight tRNA genes which were encoded on the L-strand. The phylogenetic tree of C. mccordi and 10 other closely related species was built. The DNA data presented here will be useful to study the evolutionary relationships and genetic diversity of C. mccordi.
Asian box turtles belong to the genus Cuora in the family Geoemydidae. There are 12 species which are distributed from China to Malay Peninsula, Indonesia and the Philippines, throughout mainland South-East Asia (Zhang et al. Citation1998; Spinks and Shaffer Citation2007; Spinks et al. Citation2009; Spinks et al. Citation2012). The McCord's box turtle, Cuora mccordi, endemic to China, was described by Ernst based on specimens from south-western Guangxi Zhuang Autonomous Region, China (Ernst Citation1988). After that, no individual was found in field, but some individuals are captive breeding in turtle farms or private owners (Zhou et al. Citation2008). C. mccordi was listed as data deficient level on the International Union for Conservation of Nature (IUCN) Red List in 1996, and its endangered category was set to the critically endangered level in 2000 (IUCN Citation2016). In this article, we determined and described the mitogenome of C. mccordi in order to obtain basic genetic information about this species.
The specimen of C. mccordi was a dead juvenile individual collected from a turtle farm (Shanghai City). It was preserved and deposited in the Museum of Huangshan University (Voucher numbers: HUM2015001). The complete mitogenome of C. mccordi (Genbank accession number KU258498) was sequenced to be 16,688 bp which consisted of 13 typical vertebrate protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and a control region (D-loop), which is similar to the typical mtDNA of other vertebrates (Boore Citation1999; Sorenson et al. Citation1999).
The overall base composition of the entire genome was as follows: A (34.1%), T (27.0%), C (26.1%) and G (12.8%), which the percentage of A + T (61.0%) reflected a typical sequence feature of the vertebrate mitogenome. Most of the C. mccordi mitochondrial genes are encoded on the H-strand except for the ND6 gene and eight tRNA genes, which are encoded on the L-strand. The positions of RNA genes were predicted by the MITOS (Bernt et al. Citation2013), and the locations of protein-coding genes were identified by comparing with the homologous genes of other closely related species. Among the mitochondrial protein-coding genes, the ATP8 was the shortest, while the ND5 was the longest. All the protein-coding genes initiate with ATG as start codon, except for COI gene which begins with GTG. Eight protein-coding genes end with complete stop codons (AGG and TAA), and the other five genes end with T or TA as the incomplete stop codons, which were presumably completed as TAA by post transcriptional polyadenylation (Anderson et al. Citation1981). The 22 tRNA genes range in size from 66 to 78 bp. The 12 s rRNA (965 bp) and 16 s rRNA (1604 bp), are located between the tRNA-Phe and tRNA-Leu genes and separated by the tRNA-Val gene. The control region (D-loop) of the C. mccordi mitogenome in size is 1172 bp and was located between the tRNA-Pro and tRNA-Phe genes.
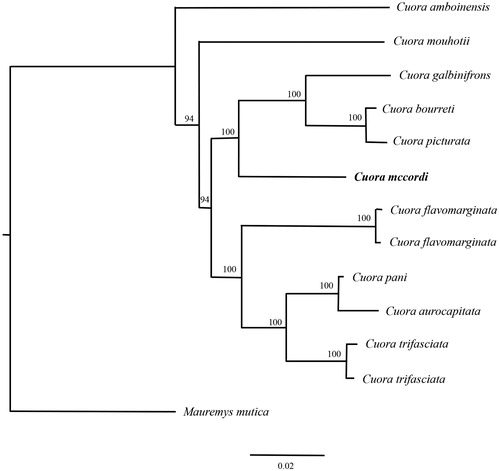
To further validate the new determined sequences, whole mitochondrial genome sequences of C. mccordi in this study, and together with other 10 related species (12 sequences) from GenBank was used to perform the phylogenetic analysis. These species were as follows: C. amboinensis, C. mouhotii, C. galbinifrons, C. bourreti, C. picturata, C. mccordi, C. flavomarginata, C. pani, C. aurocapitata, C. trifasciata, Mauremys mutica (out group). A maximum likelihood (ML) tree was constructed based on the dataset by online tool RAxML (Stamatakis et al. Citation2008). C. mccordi is the sister group to the C. galbinifrons species complex (). The phylogenetic analysis result was consistent with the previous research. It indicated that our new determined mitogenome sequences could meet the demands and explain some evolution issues.
Figure 1. A maximum likelihood (ML) tree of C. mccordi in this study and other 10 closely related species was constructed based on the dataset of the whole mitochondrial genome by online tool RAxML. The numbers above the branch meant bootstrap value. Bold black branches highlighted the study species and corresponding phylogenetic classification. The analyzed species and corresponding NCBI accession number as follows: C. amboinensis (FJ763736), C. mouhotii (DQ659152), C. galbinifrons (EU809939), C. bourreti (JN865214), C. picturata (JF712890), C. mccordi (KU258498), C. flavomarginata (EU708434, KJ680321), C. pani (GQ889364), C. aurocapitata (AY874540), C. trifasciata (KC543355, KF574821), Mauremys mutica (KP938957).
Acknowlegements
We thank Wei Lu (the owner of the turtle farm from Shanghai) for providing the specimen.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Ernst CH. 1988. Cuora mccordi, a new Chinese box turtle from Guangxi Province. Proc Biol Soc Wash. 101:466–470.
- IUCN 2016. The IUCN red list of threatened species. version 2016-3. Downloaded on 01 January. 2017. https://www.iucnredlist.org/search?query=Cuora%20mccordi&searchType=species
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. 1999. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 12:105–114.
- Spinks PQ, Shaffer HB. 2007. Conservation phylogenetics of the Asian box turtles (Geoemydidae, Cuora): mitochondrial introgression, numts, and inferences from multiple nuclear loci. Conserv Genet. 8:641–657.
- Spinks PQ, Thomson RC, Shaffer HB. 2009. A reassessment of Cuora cyclornata Blanck, McCord and Le, 2006 (Testudines, Geoemydidae) and a plea for taxonomic stability. Zootaxa. 2018:58–68.
- Spinks PQ, Thomson RC, Zhang YP, Che J, Wu Y, Shaffer HB. 2012. Species boundaries and phylogenetic relationships in the critically endangered asian box turtle genus Cuora. Mol Phylogenet Evol. 63:656–667.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 57:758–771.
- Zhang MW, Zong Y, Ma JF, et al. 1998. Fauna Sinica. Reptilia, Vol. 1. Beijing: Science Press, 100–120. (In Chinese)
- Zhou T, Blanck T, McCord WP, Li PP. 2008. Tracking Cuora mccordi Ernst, 1988: the first record of its natural habitat; a re-description; with data on captive populations and its vulnerability. Hamadryad. 32:57–69.
