Abstract
Macrobrachium rosenbergii de Man, 1879 (Caridea: Palaemonidae), commonly referred to as giant freshwater prawn, is commercially important worldwide, and a primary inland cultured species. The first complete mitochondrial genome of M. rosenbergii from China (CN) was sequenced and compared with that in Indonesia (ID) in this study. The results show that the total length of CN-mtDNA sequence is 15,767 bp (GenBank accession number: KY865098), which is shorter than that of ID-mtDNA (15,772 bp). It encodes 37 genes, including 13 protein-coding genes, 22 transfer RNA genes and 2 ribosomal RNA genes. The heavy strand and light strand encode 23 and 14 genes, respectively, and the content of A + T is 61.81%. The gene order and content of CN-mtDNA and ID-mtDNA are identical to the primitive Pancrustacean ground pattern. Phylogenetic analysis shows that CN-mtDNA is clustered into one clade with ID-mtDNA (BP =100). Pairwise genetic distance analyses indicate that the genetic distance of CN-mtDNA and ID-mtDNA is 0.086, which is smaller than values between species in the Macrobrachium genus. In summary, both phylogenetic analysis and gene re-arrrangement analysis between the M. rosenbergii in China and Indonesia have revealed that their differences are within intraspecific variation.
Macrobrachium rosenbergii de Man, 1879 (Caridea: Palaemonidae), commonly referred to as giant freshwater prawn, lives in the freshwater and brackish water habitats and is widely distributed in tropical or subtropical regions of the western Pacific Ocean (Hurwood et al. Citation2014; Maidin et al. Citation2017). Macrobrachium rosenbergii is commercially important worldwide, and is a primary inland cultured species (Cheng et al. Citation2006). The complete mitochondrial genome has been used to study the evolutionary history and phylogenetic relationships among metazoans (Boore et al. Citation1998, Citation2005; Boore and Brown Citation1998).
A specimen of M. rosenbergii was obtained from Zhejiang Institute of Freshwater Fisheries (N: 30.833, E: 120.089), China. Its mitochondrial genomic DNA was extracted and stored at Marine Museum of Huaihai Institute of Technology (Accession number: Mros-002). The results show that the total length of CN-mtDNA sequence is 15,767 bp (GenBank accession number: KY865098), which is shorter than ID-mtDNA (15,772 bp) (Miller et al. Citation2005). It encodes 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes and 2 ribosomal RNA genes. The nucleotide sequence identities in the 13 PCGs between the two M. rosenbergii range from 86.1% (atp8) to 99.5% (nd2), and the 13 PCGs together have 909 variable sites and 88 indels, with only 91.8% of invariant sites.
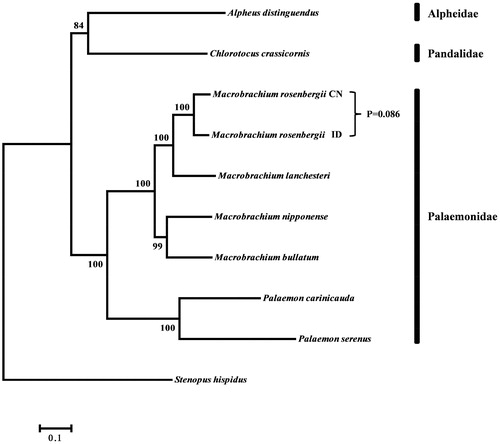
Phylogenetic tree was constructed on the basis of 13 PCGs (nucleotide sequences) of the 10 mitochondrial genomes from the infraorder Caridea (). The phylogenetic tree shows that all four genera (Chlorotocus, Alpheus, Macrobrachium and Palaemon) constitute distinct monophyletic groups, with high support value. M. rosenbergii from China is clustered into one clade with the Indonesian M. rosenbergii (BP =100). The pairwise genetic distance value between the two M. rosenbergii (CN and ID) is 0.086, which is smaller than that of other Macrobrachium species (Miller et al. Citation2005; Ma et al. Citation2011; Gan et al. Citation2015). The gene order and content of CN-mtDNA and ID-mtDNA are identical to the primitive Pancrustacean ground pattern (Miller et al. Citation2005). Both Palaemon serenus and P. carinicauda share a reversal of trnT (transfer, trn for threonine) and trnP (trn for proline) genes compared to Macrobrachium species, which retain the primitive Pancrustacean ground pattern (Shen et al. Citation2009).
Figure 1. Phylogenetic tree constructed from ML analyses based on 13 PCGs (nucleotide acid sequences) of Caridea species. The numbers at the nodes indicate the bootstrap values(BP). The accession numbers of the genomes used for comparison were NC_006880 (M. rosenbergii, ID), NC_014883 (Alpheus distinguendus), NC_012217 (Macrobrachium lanchesteri), NC_027602 (Macrobrachium bullatum), NC_015073 (Macrobrachium nipponense), NC_027601 (Palaemon serenus), NC_012566 (Palaemon carinicauda).
The natural distribution of most freshwater populations is geographically restricted. M. rosenbergii is unusual because it distributes from the Malay Peninsula, Bangladesh and India, to the Indonesian islands and West Java (Hurwood et al. Citation2014). Malaysia is the country of origin for all the currently well-known cultured strains of M. rosenbergii worldwide, so the MA population is artificially introduced into many areas (Elsheikh et al. Citation2015). The M. rosenbergii sampled from China has a few base variations, which is similar to that from Indonesia. De Jong et al. (Citation2011) attributed these subtle differences to geographical isolation. This would be in line with the reports of many authors, which have reported the genetic effects of human introduction on some freshwater animals (Shih et al. Citation2004; Wang et al. Citation2004; Liu et al. Citation2011).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL, Brown WM. 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 8:668–674.
- Boore JL, Lavrov DV, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature. 392:667–668.
- Boore JL, Macey JR, Medina M. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Meth Enzymol. 395:311–348.
- Cheng W, Tung YH, Chiou TT, Chen JC. 2006. Cloning and characterization of mitochondrial manganese superoxide dismutase (mtMnSOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 21:453–466.
- De Jong MA, Wahlberg N, Van Eijk M, Brakefield PM, Zwaan BJ. 2011. Mitochondrial DNA signature for range-wide populations of Bicyclus anynana suggests a rapid expansion from recent refugia. PLoS One. 6:e21385.
- Elsheikh MO, Mustafa FB, Eid II, Lutas A, Bhassu S. 2015. COI gene sequence analysis for testing cyclical mating in securing genetic diversity of Macrobrachium rosenbergii. Biochem Syst Ecol. 62:178–185.
- Gan HY, Gan HM, Lee YP, Austin CM. 2015. The complete mitogenome of the Australian freshwater shrimp Paratya australiensis Kemp, 1917 (Crustacea: Decapoda: Atyidae). Mitochondrial DNA Part A. 27:1–3158.
- Hurwood DA, Dammannagoda S, Krosch MN, Jung H, Salin KR, Youssef MA-BH, de Bruyn M, Mather PB. 2014. Impacts of climatic factors on evolution of molecular diversity and the natural distribution of wild stocks of the giant freshwater prawn (Macrobrachium rosenbergii). Freshwater Sci. 33:217–231.
- Liu MY, Tzeng CS, Lin HD. 2011. Phylogeography and the genetic structure of the land-locked freshwater prawn Macrobrachium asperulum (Crustacea: Decapoda: Palaemonidae) in Taiwan. Hydrobiologia. 671:1–12.
- Ma K, Feng J, Lin J, Li J. 2011. The complete mitochondrial genome of Macrobrachium nipponense. Gene. 487:160–165.
- Maidin MSR, Anton A, Yong ASK, Chin GJWL. 2017. Mitochondrial COI gene sequence of giant freshwater prawn, Macrobrachium rosenbergii: an assessment of a community-based stock enhancement programme in Petagas River, Sabah, Malaysia. Int J Fish Aquatic Stud. 5:518–526.
- Miller AD, Murphy NP, Burridge CP, Austin CM. 2005. Complete mitochondrial DNA sequences of the decapod crustaceans Pseudocarcinus gigas (Menippidae) and Macrobrachium rosenbergii (Palaemonidae). Mar Biotechnol. 7:339–349.
- Shen X, Sun MA, Wu Z, Tian M, Cheng H, Zhao F, Meng X. 2009. The complete mitochondrial genome of the ridgetail white prawn Exopalaemon carinicauda Holthuis, 1950 (Crustacean: Decapoda: Palaemonidae) revealed a novel rearrangement of tRNA genes. Gene. 437:1–8.
- Shih HT, Ng PK, Chang HW. 2004. Systematics of the genus Geothelphusa (Crustacea, Decapoda, Brachyura, Potamidae) from southern Taiwan: a molecular appraisal. Zool Stud. 43:561–570.
- Wang JP, Lin HD, Huang S, Pan CH, Chen XL, Chiang TY. 2004. Phylogeography of Varicorhinus barbatulus (Cyprinidae) in Taiwan based on nucleotide variation of mtDNA and allozymes. Mol Phylogenet Evol. 31:1143–1156.
