Abstract
Aconitum brachypodum is an endangered plant endemic to south-west China. The plastid genome of this was characterized by Illumina next-generation sequencing technology. The complete plastid genome is 155,651 bp long with a pair of 26,213 bp inverted repeat regions (IRs) that are separated by a large single copy (LSC) region of 86,292 bp and a small single copy (SSC) region of 16,933 bp. A total of 132 genes were annotated, including 85 protein-coding genes, 37 tRNA genes, eight rRNA genes and two pseudogenes. In addition, five palindromic, five forward, nine tandem repeats and 266 SSRs were detected in A. brachypodum. Furthermore, phylogenetic analysis illustrated that A. brachypodum is closely related to other ten Aconitum subgenus. Aconitum species are with high bootstrap support of 100%.
Aconitum brachypodum Diels is a perennial herb plant endemic to China and only distributed in Yunnan and Sichuan provinces, whose stem is 40–80 cm high, with blue and purple flower (Li and Kadota Citation2001). The tuberous mother roots of A. brachypodum (‘Xue-shang-yi-zhi-hao’, used by Chinese druggists) has been recorded in ‘Chinese Pharmacopoeia’ for the treatment of bruises and rheumatism (China Pharmacopoeia Committee Citation1977). Recently, with the increasing demand for wild resources, the native populations of the species have reduced dramatically. It has been listed as an endangered species in ‘China Species Red List’ (Wang and Xie Citation2004). Therefore, we need to deepen our understanding of the genetic information and raise our awareness of this species. In this study, we characterize the complete plastid genome of A. brachypodum (GenBank Accession Number: MH886505) based on the Illumina next-generation sequencing techniques.
Total genomic DNA was extracted from the fresh leaves of an individual of A. brachypodum which were collected from Yulong Snow Mountain of Yunnan (100°11′01.22″E, 27°01′25.70″N) with the modified CTAB method (Doyle and Doyle Citation1987). Voucher specimen was deposited in the herbarium of KUN. The extracted DNA was sequenced using the Illumina Miseq platform (Illumina, San Diego, CA, USA). Reads of the A. brachypodum plastid genome were assembled using CLC Genomic Workbench v10 (CLC Bio., Aarhus, Denmark). All the contigs were checked against the reference genome of A. chiisanense (NC029829) using BLAST (https://blast.ncbi.nlm.nih.gov/) and annotated using DOGMA (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004). To identify the phylogenetic position of A. brachypodum, the maximum likelihood (ML) tree was reconstructed based on 22 species complete plastid genomes by RAxML 8.2.11(Stamatakis Citation2014).
The length of A. brachypodum complete plastid genome is 155,651 bp, with a typical quadripartite structure, including a pair of inverted repeat regions (IRs) of 26,213 bp, a large single-copy (LSC) region of 86,292 bp and a small single-copy (SSC) region of 16,933 bp. It contains 132 genes, including 85 protein-coding genes, 37 tRNA genes, eight rRNA genes and two pseudogenes (ψrps19 and ψycf1). Furthermore, five palindromic, five forward and nine tandem repeats are found. The total number of SSRs is 266, of which the mono-, di-, tri-, tetra- and pentanucleotide are 130, 53, 74, 5 and 4, respectively, and no hexanucleotide is found.
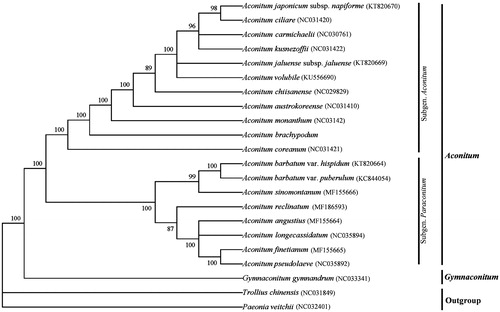
Phylogenetic analysis showed that A. brachypodum and other ten Aconitum subgenus. Aconitum species formed a monophyletic clade with 100% bootstrap support value (). Understanding the plastid genome characteristic of this species can provide valuable genetic information for further drawing up conservation strategies and conservation measures.
Acknowledgements
We appreciate the Laboratory of Molecular Biology in the Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences, for providing experimental platform.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- China Pharmacopoeia Committee. 1977. Pharmacopoeia of China. Vol.1. Beijing: People’s Medical Publishing House.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Li LQ, Kadota Y. 2001. Aconitum Linnaeus. In: Wu ZY and Raven PH editors. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 149–222.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang S, Xie Y. 2004.China Species Red List. Vol.1: Red List. Beijing: Higher Education Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.