Abstract
Ceriops tagal is an inner mangrove tree species and well known medicinal plant with wide distribution in the world mangal. Here, we reported and characterized its complete chloroplast genome sequence assembled from short reads generated by Illumina sequencing. Total chloroplast genome size was164,439 bp, including inverted repeats (IRs) of 26,390 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 92,489 bp and 20,172bp, respectively. A total of 134 genes, including 42 tRNA, 8 rRNA, and 84 protein-coding genes, were identified. Among them, 17 genes occur in IRs, containing six protein-coding genes, four rRNA genes and seven tRNA genes. The GC content of C. tagal is 35.32%. Phylogenetic analysis using the maximum likelihood algorithm showed that C. tagal is sister to a clade with three species in the order Rosales.
Ceriops tagal (Pierr.) C. B. Robinson is a long-lived, woody perennial mangrove species, which belongs to the family Rhizophoraceae and has a wide distribution (Tomlinson Citation1986). It has been proved to possess a series of antifouling terpenoids with unique structural diversity (Chen et al. Citation2008; Blunt et al. Citation2009). As an important fouling organism in East Asian coastal water (Chen et al. Citation2011), it was reported that Pimarane determinants isolated from C. tagal can significantly inhibit larval settlement of the barnacle Balanus albicostatus.
In this study, we reported and characterized the complete chloroplast genome of C. tagal using Illumina sequencing technology. The valuable data for phylogenetic inference or the population history of Ceriops can help aid in the utilization of the genetic resources of Ceriops as a medicinal mangrove tree.
Fresh leaves were collected from ten individual of C. tagal growing in Dongzhai Bay (110.32°E, 19.51°N), Hainan, China. And the total genomic DNA was extracted from ten mixed fresh leaves of C. Tagal using the modified CTAB method (Doyle Citation1987) in the laboratory of Hainan Normal University. Genome sequencing was performed on an Illumina Hiseq X Ten platform with paired-end reads of 150 bp. In total, 5.587 Mb short sequence data with Q20 was 94.88% was obtained. Low-quality reads were filtered out and the remaining high-quality reads were used to assemble the chloroplast genome in SOAPdenovo (Luo et al. Citation2012). The chloroplast rbcL and trnV sequence of C. tagal (GenBank accession number DQ145950.1 and EF423377.1) were used as the seed sequences. The genes annotation in the chloroplast genome was doing with the DOGMA Programme (Wyman et al. Citation2004). The circular chloroplast genome map was drawn by using OGDRAW and the phylogenomic tree was constructed using MAFFT version 7 (Katoh and Standley Citation2013; Lohse et al. Citation2013). Accession number on GenBank is MH240830.
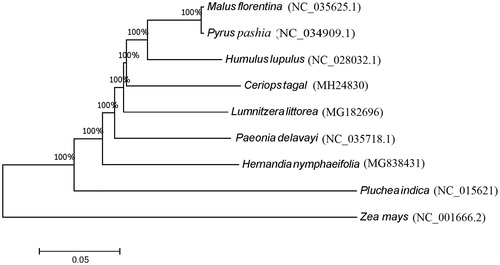
Eight chloroplast genome sequences were aligned using the complete chloroplast genomes including C. tagal and one species from Gramineae (Zea mays) was used as the outgroup species. Phylogenetic analysis using the maximum likelihood algorithm was conducted with RAxML (Stamatakis Citation2014) implemented in Geneious version 10.1 (Kearse et al. Citation2012).
A typical quadripartite structure was found in the chloroplast genome of C. tagal with a length of 164,439 bp, which contains inverted repeats (IRs) of 26,390 bp, separated by a large single copy (LSC) and a small single copy (SSC) of 92,489 bp and 20,172 bp, respectively. The chloroplast genome has a GC content of 35.32% and contains a total of 134 predicted genes, including 42 tRNAs, 8 rRNAs, and 84 protein-coding genes. Among them, 17 genes occur in IRs, containing six protein-coding genes, four rRNA genes and seven tRNA genes. Among them, 11 genes contain one intron and three genes (rps12, clpP, and ycf3) contain two introns. Phylogenetic analysis with other seven plant chloroplast genomes showed that C. tagal is sister to a clade containing three species in Rosales (). The chloroplast genome determined in this study provides useful genomic resources for characterization of genetic diversity of C. tagal.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Blunt JW, Copp BR, Hu WP, Munro MH, Northcote PT, Prinsep MR. 2009. Marine natural products. Nat Prod Rep. 26:170–244.
- Chen JD, Feng DQ, Yang ZW, Wang ZC, Qiu Y, Lin YM. 2008. Antifouling metabolites from the mangrove plant Ceriops tagal. Molecules. 13:212–219.
- Chen JD, Yi RZ, Lin YM, Feng DQ, Zhou HC, Wang ZC. 2011. Characterization of terpenoids from the root of Ceriops tagal with antifouling activity. Int J Mol Sci. 12:6517–6528.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tomlinson PB. 1986. The botany of mangroves. Q Rev Biol. 52:238.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.