Abstract
Richard’s Pipit Anthus richardi is a medium-sized passerine bird which breeds in open grasslands in northern Asia with a long-distance migrant moving to open lowlands in the Indian subcontinent and Southeast Asia. In this study, the mitochondrial genome of this bird species was obtained by the PCR-based sequencing method. The mitogenome is 16,871 bp in size, including 22 tRNAs, 2 rRNAs, 13 protein-coding genes and a D-loop. The A, T, G, and C base composition of the mitochondrial DNA are 29.73%, 23.12%, 14.59% and 32.56%, respectively. The starting codon of protein-coding genes is typical ATG, except for the ND2 which starts with ATA. The D-loop locates between genes of tRNAGlu and tRNAPhe. The result of the phylogenetic analysis indicates that A. richardi has close phylogenetic relationship with A. novaeseelandiae.
Richardʼs Pipit Anthus richardi is a medium-sized songbird from the family Motacillidae of order Passeriformes, which breeds in southern Siberia, Mongolia, parts of Central Asia and in northern, central and eastern China. It migrates south to winter in the Indian subcontinent and South-east Asia with records as far south as Sri Lanka, Singapore and northern Borneo (BirdLife International Citation2018). It was singled out from the superspecies New Zealand Pipit, A. novaeseelandiae (Hudeček Citation2017). There are five subspecies, A. r. richardi, A. r. centralasiae, A. r. dauricus, A. r. ussuriensis and A. r. sinensis (Clements et al. Citation2018). Richard’s Pipit originally belongs to A. novaeseelandiae, several subspecies of which were upgraded to A. rufulus and Anthus richardi. Understanding its mitochondrial genome structure is of great importance to the study of their systematic relationship. In this study, we obtained the complete mitochondrial genome of A. richardi, which provides insight for the study of mitochondrial genealogical genomics of passerine birds.
The mitochondrial genome of A. richardi was obtained by the PCR-based sequencing. The tissue was sampled on the 9st of September, 2010 from the dead birds which killed by bird repellent in Hefei Luogang Airport (117°18′36ʺE, 31°46′48ʺN), Anhui Province, China. It was kept in the Institute of Biodiversity and Wetland Ecology, School of Resources and Environmental Engineering, Anhui University (Sample code is ZSOF2010-01)
The complete mtDNA of A. richardi is a 16,871 bp circular molecular. The A, T, G and C base composition of the mitochondrial DNA are 29.73%, 23.12%, 14.59% and 32.56%, respectively. The content of A + T (52.85%) is higher than that in G + C (47.15%), which is similar to other Passeriformes (Yang et al. Citation2014). The mitogenome includes 22 tRNAs, 2 rRNAs, 13 protein-coding genes and a D-loop. The gene order is the same as most of other Passeriformes (Gibb et al. Citation2007). The 22 tRNA genes range from 66 bp to 79 bp. All tRNA genes possess the typical clover leaf secondary structure except for tRNASer(AGY) and tRNALeu(CUN) which lack arms of dihydrouracil (DHU) (Bernt et al. Citation2013). The mtDNA sequence consists of a small subunit (12S RNA) and a large subunit (16S RNA), separated by tRNAval. The length of 12S RNA and 16S RNA is 971 bp and 1599 bp, respectively. The starting codon of protein-coding genes is typical ATG, except for the ND2 which starts with ATA. The most of termination codon of protein-coding genes is the TAA, while COX1 and ND5 stop with AGG, COX3 and ND4 stop with T-, and ND6 stops with TAG. The D-loop is 1298 bp, located between tRNAGlu and tRNAPhe. In addition, 95 bp intergenic spacer sequences are spread over 18 regions, 7 areas of gene overlap occupy a total of 30 bp throughout the whole genome.
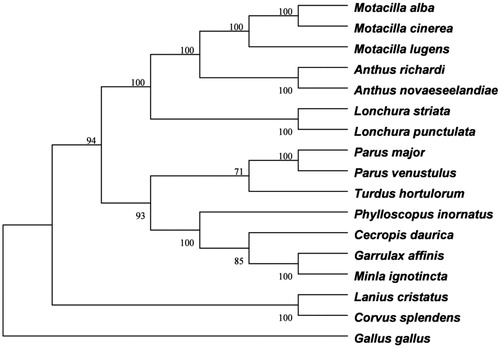
The phylogenetic tree was constructed using neighbor-joining (NJ) methods, with Gallus gallus (NC_001323) as an outgroup. The phylogenetic tree showed that Motacillidae and Ploceidae are sister groups, indicating that these two families have close phylogenetic relationship, A. richardi and A. novaeseelandiae have close relationship (). The phylogenetic relationship among Passeriformes still needs further research and more comprehensive data analysis.
Figure 1. The phylogenetic tree of 16 species Passeriformes birds constructed using the neighbour-joining method based on complete mtDNA sequences and Gallus gallus used as an outgroup. Note: The tree was built using the Kimura-2-parameter (K2P) model, and the numbers on the branches are bootstrap values. The 16 species Passeriformes included 5 Motacillidae birds: Anthus novaeseelandiae (KC545397), Motacilla alba (NC_029229), Motacilla lugens (NC_029703), Motacilla cinerea (NC_027933) and Anthus richardi (MH593382), 2 Ploceidae birds: Lonchura striata (NC_029475) and Lonchura punctulata (NC_028036), 2 Paridae birds: Parus major (NC_026293) and Parus venustulus (KP313823), 2 Muscicapidae birds: Phylloscopus inornatus (KF742677) and Minla ignotincta (KT995474), one Timaliidae bird: Garrulax affinis (KT182082), one Turdidae bird: Turdus hortulorum (KF926987), one Laniidae bird: Lanius cristatus (KT004451), one Corvidae bird: Corvus splendens (KJ766304) and one Hirundinidae bird: Cecropis daurica (KJ499911).
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Braband A, Schierwater B, Stadler PF. 2013. Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol. 69:328–338.
- BirdLife International. 2018. Anthus richardi. The IUCN Red List of Threatened Species 2016: e.T103821389A94581724. [accessed 2018 Nov 3] http://dx.doi.org/10.2305/IUCN.UK.2016-3. RLTS.T103821389A94581724.en.
- Clements JF, Schulenberg TS, Iliff MJ, Roberson D, Fredericks TA, Sullivan BL, Wood CL. 2018, The eBird/Clements checklist of birds of the world: 2018. [accessed 2018 Oct 31]. http://www.birds.cornell.edu/clementschecklist/download/.
- Gibb G, Kardailsky O, Kimball R, Braun E, Penny D. 2007. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24:269–280.
- Hudeček JJ. 2017. Historical record of Richardʼs Pipit (Anthus richardi) on the territory of Upper Silesia. Acta Mus Siles Sci Nat. 66:173–176.
- Yang F, Li B, Zhou L, Bao D, Zhu H. 2014. Complete mitochondrial genome of Tree Sparrow Passer montanus saturatus (Passeriformes: Passeridae). Mitochondrial DNA Part A. 27(3):1993–1994.
