Abstract
Aphelandra knappiae is an important plant from the family Acanthaceae. The complete chloroplast genome of the genus Aphelandra was sequenced for the first time. The plastome of A. knappiae was 152,457 bp in length. It consisted of a large single-copy (LSC) region (83861 bp) and a small single-copy (SSC) region (17,888 bp), which were separated by two inverted repeats (IRs, 25,354 bp). This plastome contained 114 unique genes, including 80 protein-coding genes, 30 tRNAgenes, and 4 rRNA genes. The overall GC content was 38.5%. Furthermore, phylogenetic analysis of five species in the Acanthaceae was also conducted. The whole chloroplast genome of this species will be useful for the future plant evolutionary, phylogeny and genomic studies in the family Acanthaceae.
Aphelandra R. Brown is a neotropical genus of Acanthaceae with about 230 species. (Daniel Citation1991). Aphelandra knappiae is an important plant described in 2013 (Wasshausen Citation2013). It has beautiful yellow flowers, which made it a potential landscaping plant. In this study, the complete chloroplast genome of the genus Aphelandra was sequenced for the first time. The study will accordingly facilitate our understanding of the chloroplast genome feature of Acanthaceae.
The silica-gel dried leaves of A. knappiae were collected from Peru, Province Rioja, District Pardo Miguel, Bosque de Proteccion Alto Mayo, on the way from Agues Verdes (5°40'49.48"S, 77°37'47.50"W, 1150 m) cross stream to (5°40'07.53"S, 77°38'22.95"W, 1015 m) Playa Azu (5°39'24.84"S, 77°38'50.84"W, 1268 m). A voucher specimen was deposited at the South China Botanical Garden Herbarium, Guangzhou, China. Total genomic DNA was isolated with a modified CTAB method (Doyle and Doyle Citation1987). Pair-end (PE) sequencing was performed on the Illumina HiSeq 2500 instruments. The sequenced clean PE reads were filtered using GetOrganelle pipeline (Jin et al. Citation2018) to get plastid-like reads. The filtered reads were assembled using SPAdes version 3.11.1 (Bankevich et al. Citation2012). To retain pure chloroplast contigs, the final ‘fastg’ files were filtered using the ‘slim’ script of GetOrganelle. The filtered De Brujin graphs were viewed and edited using Bandage (Wick et al. Citation2015). Then a circular chloroplast genome was generated. The genome was automatically annotated using Plastid Genome Annotator (PGA) (https://github.com/quxiaojian/PGA), coupled with manual correction of start and stop codons and intron/exon boundaries using Geneious version 11.0.4 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). The final complete plastome was deposited into GenBank (accession number MH909777).
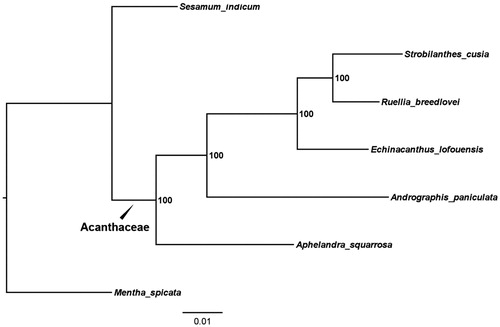
To reconstruct the phylogeny of Acanthaceae, Five Acanthaceae cp genomes were included in the phylogenetic analysis. The published complete genomes were downloaded from the NCBI GenBank database, including Andrographis paniculata (KF_150644), Echinacanthus lofouensis (MF_490441), Ruellia breedlovei (KP_300014), Strobilanthes cusia (MG_874806), Mentha spicata (NC_037247), and Sesamum indicum (NC_016433) were hired as outgroups. The plastomes (each excluding one IR) of all the species were aligned using MAFFT version 1.3.7 (Katoh and Standley Citation2013) implemented in Geneious version 11.0.4. The maximum likelihood (ML) phylogeny was reconstructed using RAxML version 8.0.0 (Stamatakis Citation2014).
The complete cp genome of A. knappiae was 152,457 bp in length and presented a typical quadripartite structure including a large single copy (LSC) region (83,861 bp), a small single copy (SSC) region (17,888 bp), and two inverted repeat regions (IRs) (25,354 bp). The overall GC content of A. knappiae plastome was 38.5%. The A. knappiae plastome contained 114 different genes, including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
The phylogeny analysis recovered three fully supported monophyletic lineages (bootstrap = 100%) in the family Acanthaceae, which correspond to the study by McDade et al. (Citation2008) (). This complete chloroplast genome can be subsequently used for population, phylogenetic and cp genomic studies of the family Acanthaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Daniel TF. 1991. A revision of Aphelandra (Acanthaceae) in Mexico. Proc Calif Acad Sci. 47:235–274.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- McDade LA, Daniel TF, Kiel CA. 2008. Toward a comprehensive understanding of phylogenetic relationships among lineages of Acanthaceae sl (Lamiales). Am J Bot. 95:1136–1152.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wasshausen DC. 2013. New species of Aphelandra (Acanthaceae) from Peru and Ecuador. J Bot Res Inst Tex. 7:109–120.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.