Abstract
Beaked whales of the genus Mesoplodon are rarely encountered and information on their basic biology is lacking. We sequenced the complete mitochondrial genomes of True’s (Mesoplodon mirus) and Sowerby’s (Mesoplodon bidens) beaked whales to high depth (>120X). The length of the de novo assembled genomes was 16,342 bp (M. mirus) and 16,347 bp (M. bidens), with GC compositions of 37.7 and 39.2%, respectively. A maximum-likelihood phylogeny supports the hypotheses that M. mirus is most closely related to Mesoplodon europaeus, and that M. bidens is the basal species of the genus. These data provide molecular resources that will aid in further understanding population genetics and evolution in this rare and poorly understood group of cetaceans.
The genus Mesoplodon is the most speciose genus in the order Cetacea (Wilson Citation1992), yet its 15 species described to date are some of the most poorly known of all large mammals (Ellis and Mead Citation2017; Jefferson et al. Citation2015). All members of the genus Mesoplodon are listed as ‘Data deficient’ by the International Union for Conservation of Nature and Natural Resources Red List (Taylor et al. Citation2008). In Canada, Sowerby’s beaked whales are listed under the national Species at Risk Act as ‘Special Concern’ due to anthropogenic threats (Fisheries and Oceans Canada Citation2017). Existing genetic resources for mesoplodonts are limited, with mitogenomes available for only five species (Thompson et al. Citation2016; Lee et al. Citation2017). Here, we describe the first complete mitogenomes for True’s and Sowerby’s beaked whales, Mesoplodon mirus and Mesoplodon bidens.
Samples were collected from dead beach cast animals by local stranding networks: muscle tissue of an adult female M. bidens in North Sydney (Nova Scotia, Canada; 46°12'13.6''N, 60°14'39.8''W) in 2016, and skin tissue from a juvenile female M. mirus in the Îsles de la Madeleine (Québec, Canada; 47°40'37.5''N, 61°29' 18.5''W) in 2017. Tissue and DNA are stored in the Marine Gene Probes laboratory at Dalhousie University. Sex was determined by presence or absence of the uterus during necropsies. Genomic DNA was extracted using phenol-chloroform for M. bidens (Sambrook and Russell Citation2006) and glass milk for M. mirus (modified from Elphinstone et al. Citation2003). Libraries were prepared using Nextera Kits with unique index adapters (M. bidens: i5-05, i7-04; M. mirus: i5-05, i7-12) and sequenced on Illumina HiSeq at The Center for Applied Genomics (Toronto, Canada). We demultiplexed using bcl2fastq2 (Illumina), trimmed using Trimmomatic (Bolger et al. Citation2014), and filtered by alignment to the Hyperoodon ampullatus mitogenome using Bowtie2 (Langmead and Salzberg Citation2012). Reads were assembled de novo using MIRA-v4.0.2 (Chevreux et al. Citation1999), corrected for errors using Pilon (Walker et al. Citation2014), and annotated with MITOS2 (Bernt et al. Citation2013).
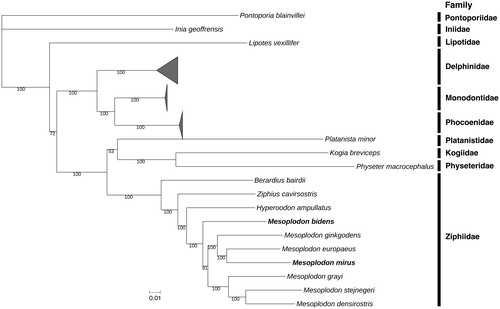
The circular mitogenomes of M. bidens (MH922777; 120× coverage) and M. mirus (MH922776; 166× coverage) are respectively 16,347 and 16,342 bp in length, with overall GC compositions of 39.2 and 37.7%. Both have typical vertebrate mitochondrial features, including 22 transfer RNA genes, 13 protein-coding genes, and two ribosomal RNA genes. We created a phylogenetic tree using mitogenomes of M. bidens, M. mirus, and 38 other toothed whales (Odontoceti). All sequences were aligned using Clustal Omega (Sievers et al. Citation2011), with 2.31% of the multiple sequence alignment containing gaps. We inferred maximum likelihood phylogenetic relationships with RAxML v8.2.9 (Stamatakis Citation2006) using a GTR substitution matrix and branch support assessed with 1000 bootstrap replicates. The tree () suggests that M. bidens is the basal species of the Mesoplodon genus, and that M. mirus is most closely related to Mesoplodon europaeus. These results are consistent with phylogenetic tree topology of Mesoplodon using seven nuclear introns (Dalebout et al. Citation2008), indicating evolutionary concordance between nuclear and mitochondrial markers.
Acknowledgements
The tissue sample of M. bidens was provided by the New Brunswick Museum and collected by the Nova Scotia Marine Animal Response Society. The M. mirus specimen was collected and provided by the Québec Marine Mammal Emergency Response Network.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol. 99:45–56.
- Dalebout ML, Steel D, Baker CS. 2008. Phylogeny of the beaked whale genus Mesoplodon (Ziphiidae: Cetacea) revealed by nuclear introns: implications for the evolution of male tusks. Syst Biol. 57:857–875.
- Ellis R, Mead JG. 2017. Beaked whales: a complete guide to their biology and conservation. Baltimore (MD): JHU Press.
- Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ. 2003. An inexpensive and high‐throughput procedure to extract and purify total genomic DNA for population studies. Mol Ecol Notes. 2:317–320.
- Fisheries and Oceans Canada. 2017. Management plan for the Sowerby's beaked whale (Mesoplodon bidens) in Canada. Species at risk act management plan series. Canada, Ottawa: Fisheries and Oceans; p. 46.
- Jefferson TA, Pitman RL, Webber MA. 2015. Marine mammals of the world. 2nd ed. Amsterdam: Elsevier.
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Lee K, Lee J, Cho Y, Kim HW, Park KJ, Sohn H, Choi YM, Kim HK, Jeong DG, Kim JH. 2017. Characterization of the complete mitochondrial genome of Stejneger’s beaked whale, Mesoplodon stejnegeri (Cetacea: Ziphiidae). Conserv Genet Resour. 839–842.
- Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol: chloroform. CSH Protoc. 1:pdb-rot4455.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 7:539.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Taylor BL, Baird R, Barlow J, Dawson SM, Ford J, Mead JG, Notarbartolo di Sciara G, Wade P, Pitman RL. 2008. Mesoplodon bidens, M. bowdoini, M. carlhubbsi, M. densirostris, M. europaeus, M. ginkgodens, M. grayi, M. hectori, M. layardii, M. mirus, M. perrini, M. peruvianus, M. stejnegeri, M. traversii. The IUCN red list of threatened species 2008. http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T13250A3430702.en. Last accessed: November 3, 2018.
- Thompson KF, Patel S, Williams L, Tsai P, Constantine R, Baker CS, Millar CD. 2016. High coverage of the complete mitochondrial genome of the rare Gray’s beaked whale (Mesoplodon grayi) using Illumina next generation sequencing. Mitochondrial DNA A. 27:128–129.
- Wilson EO. 1992. Diversity of life. Cambridge (MA): Harvard University Press.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963.