Abstract
The Brachymystax tsinlingensis Li (Salmoniformes, Salmonidae) is a freshwater resident salmonid fish. In this study, the complete mitochondrial genome of B. tsinlingensis was determined. The mitochondrial genome is 16,832 bp in size, including 22 tRNA genes, 13 protein-coding genes, 12S rRNA, 16S rRNA and one control region. The gene order of B. tsinlingensis was similar to those of most other vertebrates. The overall base composition of B. tsinlingensis is 28.3% A, 26.7% T, 16.3% G and 28.7% C, with a slight AT rich feature (55.0%). Phylogenetic analyses showed that B. tsinlingensis has a close genetic relationship to B. tumensis.
Brachymystax tsinlingensis Li is a freshwater resident salmonids distributed in streams and rivulets of Qinling Mountains of China. It belongs to the genus Brachymystax of the subfamily Salmoninae within the Salmonidae, the genus Brachymystax is comprised of B. lenok, B. tsinlingensis and B. tumensis. There are many taxonomic problems due to phenotypic similarities within the genus Brachymystax. Previously, B. tsinlingensis was described as B. lenok tsinlingensis, an endemic subspecies and considered the Brachymystax fishes distributed in Amur River, Yalu River and Tumen River of the nominate subspecies, named as B. lenok lenok (Pallas) (Li Citation1966). Recently, researchers have compared three lenok-groups, B. lenok, B. tumensis and B. lenok tsinlingensis, based on the morphological results (Xing et al. Citation2015; Meng et al. Citation2018a; Meng et al. Citation2018b) and the mitochondrial DNA fragments (i.e. cytb, part of D-loop region) (Xing et al. Citation2015; Du et al. Citation2016), and then, they recommended that the B. lenok tsinlingensis has reached species level (Xing et al. Citation2015; Meng et al. Citation2018b), so we use the latin name B. tsinlingensis in the present study. As the blurry status of B. tsinlingensis, it is necessary to obtain more evidence, such as the whole mitochondrial genome sequence of B. tsinlingensis, to contribute to the species validation, and mitochondrial genes are most commonly used markers in species identification. Furthermore, this work can also provide some available information for clarifying the genetic relationship of genus Brachymystax.
In this study, we sequenced the complete mitochondrial genome of B. tsinlingensis using a small amount of muscle from the body of fresh fish with the GenBank Accession No. KY124134. The latitude and longitude of sampling location is 34°02′12″ N, 107°37′06″ E and the DNA were stored in Key Laboratory of Animal Genetics and Breeding, Northwest A and F University. Primers were designed according to the mitochondrial genome sequence of B. tumensis. The complete genome is 16 822 bp in size and contains 22 tRNA genes, 13 protein-coding genes (PCGs), 12SrRNA, 16S rRNA and one control region. The overall base composition of B. tsinlingensis is 28.3% A, 26.7% T, 16.3% G and 28.7% C, with a slight AT rich feature (55.0%). The gene content, arrangement, and base composition of B. tsinlingensis are almost identical to that found in most teleost mitochondrial genomes (Miya and Nishida Citation1999; Broughton et al. Citation2001; Cui et al. Citation2009; Wang et al. Citation2011). The heavy DNA strand (H-strand) carried most of the genes: 12 protein-coding genes, two rRNA and 14 tRNA. ND6 and eight tRNA genes were encoded on the L-strand. The lengths of 22 tRNA genes ranged from 67 to 75 bp, and all of them could fold into the typical cloverleaf-shaped secondary structure. The non-coding regions, L-strand replication origin and control region, were 34 and 1175 bp in length, respectively. The total length of the 13 protein-coding gene was 11 422 bp, which accounts for 67.9% of the mitochondrial genome. 10 intergenic spacer (ranging from 1 to 14 bp) and eight overlap spacer (range from 1 to 10 bp) were found in whole mitochondrial genome.
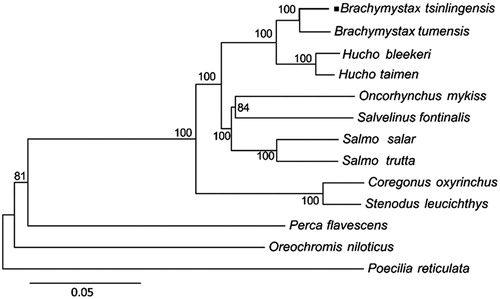
The mitochondrial genome has been broadly applied in population genetics and phylogenetic (Singh Citation2008; Galtier et al. Citation2009). To further validate the phylogenetic relationships within genus Brachymystax, phylogenetic analyses of 13 fishes were conducted through using all aligned positions of gene-coding regions of mitogenomic sequences. A neighbour-joining (NJ) tree was constructed using MEGA 6.0 (Tamura et al. Citation2013). The phylogenetic tree showed that B tsinlingensis and B. tumensis formed a monophyletic group of genus Brachymystax with high bootstrap support values (100%) (), genus Hucho was the most closed group with genus Brachymystax in the phylogenetic tree. Further study could be conducted to develop and characterize SNP markers to compare the three of B. lenok, B. tsinlingensis and B. tumensis by RAD-sequencing in phylogenetic analysis and Brachymystax species identification.
Acknowledgements
We gratefully acknowledge S. E. Wen, H. B. Sheng and P. Li for their help during the experiment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11:1958–1967.
- Cui Z, Liu Y, Li CP, You F, Chu KH. 2009. The complete mitochondrial genome of the large yellow croaker, Larimichthys crocea (Perciformes, Sciaenidae): unusual features of its control region and the phylogenetic position of the Sciaenidae. Gene. 432:33–43.
- Du Y, Wang T, Yang S, Shi X. 2016. Discussion on species validity of Brachymystax lenok tsinlingensis in Qinling Mountains. Gansu Anim Veter Med. 46:122–126. [In Chinese]
- Galtier N, Nabholz B, Glémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541–4550.
- Li S. 1966. On a new subspecies of fresh-water trout, Branchymystax lenok tsinlingensis, from Taibaishan, Shannxi, China. Acta Zootaxon Sinica. 3:92–94. [In Chinese]
- Meng Y, Wang G, Xiong D, Liu H, Liu X, Wang L, Zhang J. 2018a. Geometric morphometric analysis of the morphological variation among three lenoks of genus Brachymystax in China. Pak J Zool. 50:885–895.
- Meng Y, Wang G, Xiong D, Liu H, Zhang J, Wang J, Wang L, Liu X. 2018b. The validity of Brachymystax lenok tsinlingensis Li based on morphological difference analysis. Acta Hydrobiologica Sinica. 42:553–563. [In Chinese]
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Marine Biotechnol. 1:416–426.
- Singh TR. 2008. Mitochondrial gene rearrangements: new paradigm in the evolutionary biology and systematics. Bioinformation. 3:95–97.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang Y, Zhang XY, Yang SY, Song ZB. 2011. The complete mitochondrial genome of the taimen, Hucho taimen, and its unusual features in the control region. Mitochondrial DNA. 22:111–119.
- Xing YC, Lv BB, Ye EQ, Fan EY, Li SY, Wang LX, Zhang CG, Zhao YH. 2015. Revalidation and redescription of Brachymystax tsinlingensis Li, 1966 (Salmoniformes: Salmonidae) from China. Zootaxa. 3962:191–205.