Abstract
We have sequenced the complete mitochondrial genome (mitogenome) of the leafhopper Evacanthus heimianus in Evacanthinae, the mitogenome is 15,806 bp, has an A + T content of 79.89% (A 40.7%; T 39.2% C 10.7%; G 9.4%), which is the classical structure for insect mitogenome. All PCGs started with ATN and stopped with TAN, except ATP8 and ATP6, which started with TTG. The phylogenetic tree confirms that E. heimianus and Sophonia linealis are clustered into a clade. This study enriches the mitogenomes of the Evacanthinae subfamily.
Evacanthus heimianus (Kuoh 1980) is a species of Evacanthinae subfamily that is widely and abundantly distributed in China (Li and Wang Citation1994). Previously, only one partial mitochondrial genome (mitogenome) of Evacanthinae had been sequenced and reported (Sophonia linealis, KX437723) (Song et al. Citation2018).
In this study, the complete mitogenome sequences of Evacanthus heimianus were determined using next-generation sequencing method for the first time, which obtained for leafhoppers in this study will facilitate future studies on the identification, population genetics, and evolution of Evacanthinae leafhoppers.
Total genome DNA was extracted from male adult of E. heimianus which was collected form Sichuan Province (101°46′E, 30°10′N), China in August 2016. And voucher specimen’s genome DNA and male external genitalia are deposited in the Institute of Entomology, Guizhou University, Guizhou Province, China. The complete mitogenome of E. heimianus is 15,806 bp in length (GenBank accession number MG813486), containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one large non-coding region (Control region), and two kinds of repeat units were founded in control region. In general, the E. heimianus mitogenome has an A + T content of 79.89% (A 40.7%; T 39.2% C 10.7%; G 9.4%), which is within the range reported from Hemipteran mitogenomes (68.86–86.33; Wang et al. Citation2015). All PCGs started with ATN and stopped with TAN, except ATP8 and ATP6, which started with TTG. All tRNA genes are identified by ARWEN version 1.2 software (Laslett and Canbäck Citation2008). The 16S rRNA gene is 1170 bp in size and is located between tRNA-L2 and tRNA-V; the 12S rRNA gene is 733 bp in length and is located after tRNA-V. The control region is 1,549 bp long, and is located between 12S rRNA and tRNA-I.
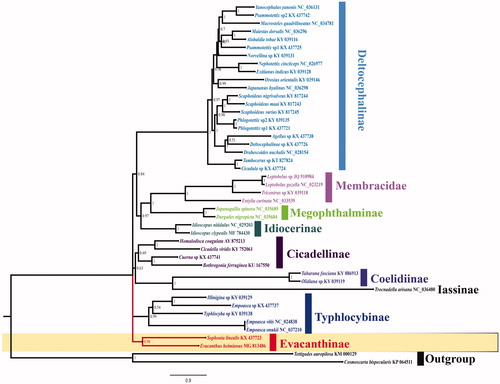
The phylogenetic relationships of the E. heimianus were reconstructed using Bayesian analysis based upon the concatenated nucleotide sequences of the 13 PCGs (). Sequences were aligned using MEGA7 software (Kumar et al. Citation2016). Phylogenetic trees were generated using PhyloBayes software (Miller et al. Citation2010) with the GTR + CAT model. The phylogenetic tree confirms that E. heimianus is part of the Evacanthinae, all members of which are clustered into a clade. Up to now, few studies have been recorded for Evacanthinae, and we hope that our data can useful for further study.
Figure 1. Phylogenetic analyses of Evacanthus heimianus based upon the concatenated nucleotide sequences of the 13 PCGs of 44 species. The analysis was performed using PhyloBayes software. Numbers at nodes are bootstrap values. The accession number for each species is indicated after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24: 172–175.
- Li ZZ, Wang LM. 1994. A new genus and three new species of the tribe Evacanthini (Insecta: Homoptera: Cicadellidae) with a key to the genera and a list of species occurring in China. J Natural Hist. 28:373–382.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees in Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA pp 1–8.
- Song N, Cai WZ, Li H. 2018. Insufficient power of mitogenomic data in resolving the auchenorrhynchan monophyly. Zool J Linnean Soc. 183: 776–790.
- Wang Y, Chen J, Jiang LY, Qiao GX. 2015. Hemipteran mitochondrial genomes: features, structures and implications for phylogeny. Int J Mol Sci. 16:12382–12404.
