Abstract
The complete nucleotide sequence of Pinus sibirica Du Tour chloroplast genome (cp DNA) was determined in this study. The cpDNA was 116,635 bp in length, containing a pair of 473 bp inverted repeat (IR) regions (IRa and IRb), which were separated by large and small single-copy regions (LSC and SSC) of 63,908 and 51,781 bp, respectively. The cpDNA contained 113 genes, including 81 protein-coding genes, 4 ribosomal RNA genes, and 28 tRNA genes. The majority of these genes were single copy genes, while 4 genes existed as double copies, including 2 protein-coding genes (psbA and ycf2), 2 tRNA genes (trnAP and trnS). The overall GC content of P. sibirica cpDNA is 38.7%, while the corresponding values of the LSC, SSC, and IR regions are 38.0, 39.7, and 37.8%, respectively. The phylogenetic analysis confirmed clusterization of P. sibirica in Strobus subgenus.
Keywords:
Siberian stone pine (Pinus sibirica Du Tour) is a boreal species of forest woody plants with a vast mostly Siberian area of more than 5 million km2. It is widespread from the North-East of the Russian plain in the West to the Aldan upland in the East, from the Arctic Circle in Northern Siberia to the Hangay Mountains in Mongolia. In this study, we assemble and characterize the complete chloroplast genome of Pinus sibirica Du Tour. The annotated cpDNA of P. sibirica has been deposited in the GenBank with the accession number NC_028421 (KT723438).
Shoots of P. sibirica were collected near Abaza (Russia, the Western Sayan, 52°65′N, 90°01′E), grafted at (local) seedlings and cultivated at “Kedr” field station (scientific collection: 507474) (Russia, South of Tomsk, 56°13′N, 84°51′E), managed by the Institute of Monitoring of Climatic and Ecological Systems SB RAS. Genomic DNA was isolated from fresh needles using a PowerPlant Pro DNA Isolation Kit (Mo-Bio, Carlsbad, CA). The whole genome was sequenced on the MiSeq platform (Illumina) in SB RAS Genomics Core Facility (ICBFM SB RAS, Novosibirsk, Russia). The chloroplast genome was assembled using CLC GW version 8.0 (CLC Bio, Aarhus, Denmark). The gene annotations were performed using DOGMA (Wyman et al. Citation2004).
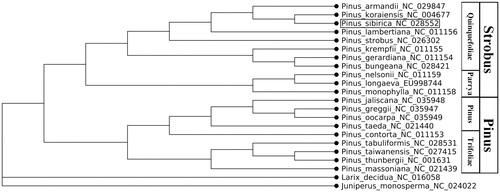
For the 20 conifers species with known whole chloroplast genomes, we used the nucleotide sequences and aligned them by MAFFT version 7.407 (Katoh and Standley Citation2013) with default parameters. Sequences from Larix decidua and Juniperus monosperma were used as an outgroup. Phylogenetic trees were inferred from the alignments in MEGA version 10.0.4 (Kumar et al. Citation2018). Maximum likelihood (ML) tree construction methods were used with 1000 bootstrap re-sampling. The GTR + G substitution model was used for ML analysis.
P. sibirica, the circular double-stranded cpDNA sequence of 116,635 bp consists of two inverted repeat (IR) regions of 473 bp each, a large single-copy (LSC) region of 63,908 bp and a small single-copy (SSC) region of 51,781 bp. The overall base composition of the cp genome in asymmetric order is A: 30.6%, C: 19.4%, G: 19.3%, and T: 30.7%, and the AT content is 61.3% and GC content 38.7%. The proportions of AT contents in LSC, SSC, and IR regions are 62.0, 60.3, and 62.0%, respectively. Using the online programme DOGMA, a total of 87 genes were identified in the cp genome, including 81 protein-coding genes, 28 tRNAs, and 4 rRNAs. The majority of these genes were single copy genes, whereas 6 genes existed as double copies, including 2 protein-coding genes psbA and ycf2), 2 tRNA genes (trnAP and trnS). A phylogenetic analysis of 20 whole chloroplast genomes from the genus Pinus is consistent with classification (Gernandt et al. Citation2005). P. sibirica is expectedly located in section Quinquefoliae of subgenus Strobus ().
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Gernandt DS, Geada López G, Ortiz García S, Liston A. 2005. Phylogeny and classification of Pinus. Taxon. 54:29–42.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.