Abstract
Pristicon trimaculatus is a tropical reef-associated marine fish belonging to the family Apogonidae. Herein, we report the first sequencing and assembly of the complete mitochondrial genome of P. trimaculatus. The complete mitochondrial genome is 16,512 bp length and has the typical vertebrate mitochondrial gene arrangement, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. Phylogenetic analysis using mitochondrial genomes of nine species showed that P. trimaculatus formed a monophyletic group with other Apogonidae species. This mitochondrial genome provides potentially important resources for addressing taxonomic issues of the Kurtiformes.
The three-spot cardinalfish, Pristicon trimaculatus (Kurtiformes, Apogonidae), is a tropical marine fish which inhabits inshore coral reefs of the western Pacific ocean, from Ryukyu Islands to Western Australia and the southern Great Barrier Reef, east to Samoa and the Marshall Islands (Randall and Fraser 1999). Its belonging family Apogonidae, the cardinalfishes, are distinguished by their large mouths and the division of the dorsal fin into two separate fins (Johnson et al. Citation1998). This species is also known to broods its eggs inside the mouth (Thresher Citation1984). In numerous molecular studies, Apogonidae, Gobiodei and Kurtus formed monophyletic group (Li et al. Citation2009; Thacker 2009; Near et al. Citation2012; Near and Keck Citation2013; Betancur-R et al. Citation2014), but not strongly supported in the bootstrap analysis (Thacker et al. Citation2015). This is the first study to determine the complete mitochondrial genome of P. trimaculatus, and to analyze the phylogenetic relationship of this species with members of Apogonidae.
The P. trimaculatus specimen was collected from Chuuk ST 1, Micronesia (7.27N, 151.54E). Total genomic DNA was extracted from tissue of the specimen, which has been deposited in the National Marine Biodiversity Institute of Korea (Voucher No. MABIK 0000642). The mitogenome was sequenced and assembled using Illumina Hiseq 4000 sequencing platform (Illumina, San Diego, CA) and SOAPdenovo assembler at Macrogen Inc. (Korea), respectively. The complete mitochondrial genome was annotated using MacClade ver. 4.08 (Maddison and Maddison Citation2005) and DNASIS ver 3.2 (Hitachi Software Engineering, Tokyo, Japan).
The complete mitochondrial genome of P. trimaculatus (GenBank accession no. AP018928) is 16,512 bp length, and includes 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. The ND6 gene and eight tRNA genes are encoded on the light strand. The overall base composition of the heavy strand is 27.68% A, 29.19% C, 16.57% G, and 26.56% T. Similar to the mitogenomes of other vertebrates, the AT content is higher than the GC content (Saccone et al. Citation1999). All tRNA genes can fold into a typical cloverleaf structure, with lengths ranging from 66 to 74 bp. The 12S rRNA (957 bp) and 16S rRNA genes (1685 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 12 begin with an ATG start codon; the exception being the COI gene, which start with GTG. The stop codon of the protein-coding genes is TAA in ND1, ND2, COI, ATP8, ND4L and ND5; TA in ATP6 and COIII; AGA in ND6; and T in the remaining four genes. The control region (844 bp) is located between tRNAPro and tRNAPhe.
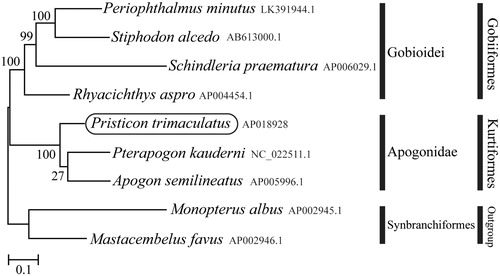
Phylogenetic trees were constructed by the maximum-likelihood method using MEGA 7.0 software (Kumar et al. Citation2016) for the newly sequenced genome and a further nine complete mitochondrial genome sequences downloaded from the National Center for Biotechnology Information. We confirmed that P. trimaculatus formed a monophyletic group with other Apogonidae species with high statistical support (). This mitochondrial genome provides important resources for addressing taxonomic issues and studying molecular evolution.
Disclosure statement
The authors report no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Betancur-R R, Wiley EO, Miya M, Lecointre G, Bailly N, Ortí G. 2014. New and revised classification of bony fishes. Version 2. <http://www.deepfin.org/Classification_v2.htm> (Electronic version accessed 24.06.14).
- Johnson GD, Gill AC, Paxton JR, Eschmeyer WN. 1998. Encyclopedia of Fishes. San Diego (CA): Academic Press; p. 183.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li B, Dettaï A, Cruaud C, Couloux A, Desoutter-Meniger M, Lecointre G. 2009. RNF213, a new nuclear marker for acanthomorph phylogeny. Mol Phylogenet Evol. 50:345–363.
- Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. ver. 4.08. Sunderland (MA): Sinauer Associates.
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad Sci USA. 109:13698–13703.
- Near TJ, Keck BP. 2013. Free from mitochondrial DNA: nuclear genes and the inference of species trees among closely related darter lineages (Teleostei: Percidae: Etheostomatinae). Mol Phylogenet Evol. 66:868–876.
- Randall JE, Fraser TH. 1999. Clarification of the western Pacific cardinalfish species Apogon trimaculatus and A. rhodopterus, with description of a similar new species. Raffles Bull Zool. 47:617–633.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.
- Thacker CE. 2009. Phylogeny of Gobioidei and placement within Acanthomorpha, with a new classification and investigation of diversification and character evolution. Copeia. 2009:93–104.
- Thacker CE, Satoh TP, Katayama E, Harrington RC, Eytan RI, Near TJ. 2015. Molecular phylogeny of Percomorpha resolves Trichonotus as the sister lineage to Gobioidei (Teleostei: Gobiiformes) and confirms the polyphyly of Trachinoidei. Mol Phylogenet Evol. 93:172–179.
- Thresher RE. 1984. Reproduction in reef fishes. Neptune City (NJ): T.F.H. Publications; p. 399.