Abstract
The complete mitochondrial genome of Epeus alboguttatus (Araneae: Salticidae) is 14,625 bp in length (GenBank accession number MH922026). It was predicted to consist of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes, and a putative control region. The gene composition and orientation of E. alboguttatus were identical to those found in other spider mitogenomes. The nucleotide composition shows heavily biased towards As and Ts, accounting for 77.59%. Twenty-two genes were in majority strand and 15 genes were in minority strand. ATT, ATA, TTA, and TTG were initiation codons, and TAA, TAG, and T were termination codons. Twenty-two reading frame overlaps and four intergenic regions were found in the mitogenome of E. alboguttatus. Ten tRNA genes lost the TΨC arm stems and had unpaired amino acid acceptor arms, whereas three tRNAs lacked the dihydrouracil (DHU) arm. Phylogenetic analysis indicated that E. alboguttatus is nested with Salticidae and recovered as sister to other five salticids.
The jumping spider belongs to the family Salticidae, a large family in Araneae, having about 5000 species in worldwide (Maddison and Hedin Citation2003; Maddison et al. Citation2008). The species, Epeus alboguttatus (Thorell, 1887) (Araneae: Salticidae) owning a varied body colour and has been found in Burma, Vietnam, and China. In wild field, this species inhibits in low altitude forests or grass. In this study, adult specimens of E. alboguttatus were collected from Libo county, Guizhou Province, China (N25°18′, E107°56′), and were stored in the spider specimen room of Guiyang University with an accession number GYU-GZML-33.
The complete mitogenome of E. alboguttatus (GenBank accession number MH922026) is a typical circular duplex DNA molecule with a length of 14,625 bp. It was predicted to consist of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS), and a putative control region that are typically present in metazoan mitogenomes (Boore Citation1999). The gene composition and orientation of E. alboguttatus were identical to those found in other spider mitogenomes (Pan et al. Citation2014; Zhu and Zhang Citation2017). Among these 37 genes, 22 genes were coded on the major strand (J-strand) while the others were coded on the minor strand (N-strand). The nucleotide composition of E. alboguttatus mitogenome shows heavily biased towards As and Ts (A, G, C, and T was 36.76%, 13.88%, 8.53%, and 40.83%, respectively), with A + T contents of 77.59%. The AT-skew and GC-skew of this genome were −0.052 and 0.239, respectively.
The E. alboguttatus mitogenome has a total of 91 bp intergenic spacer sequences, which is made up of 7 regions in the range from 3 to 74 bp. The longest intergenic spacer of 74 bp is located between trnN and trnA. Gene overlaps were found in 22 gene junctions and involved a total of 213 bp. The longest overlap is 32 bp in length and located between nad3 and trnL2. The control region was located between trnQ and trnM genes with a length of 968 bp, and the A + T content was 82.64%. The rrnL was located between trnL1 and trnV, and rrnS resided between trnV and trnQ, respectively. The rrnL was 1006 bp in length with A + T content of 81.61%, and the rrnS was 696 bp in length with A + T content of 80.89%.
The length of 22 tRNAs ranged from 50 bp (trnR) to 65 bp (trnG, trnV, and trnQ), A + T content ranged from 62.75% (trnS1) to 87.27% (trnS2). The standard cloverleaf secondary structure was found in 9 tRNA genes. Ten of the tRNA sequences (trnM, trnW, trnK, trnD, trnR, trnE, trnF, trnH, trnP, and trnT) lack the potential to form TΨC-stem, three tRNAs (trnA, trnS1, and trnS2) lost the dihydrouracil (DHU) arm and simplified down to a loop. The cox1 gene begin with TTA as start codon, cox2, nad4, and cox3 genes begin with TTG as start codon, nad2, atp8, nad4L, and cob genes begin with ATT as start codon, and other 5 PCGs start with ATA. Eight PCGs including cox1, cox2, cox3, atp6, atp8, nad3, nad6, and cob are terminated with TAA as stop codon, nad2, nad4, nad4L, and nad5 end with T, and nad1 ends with TAG.
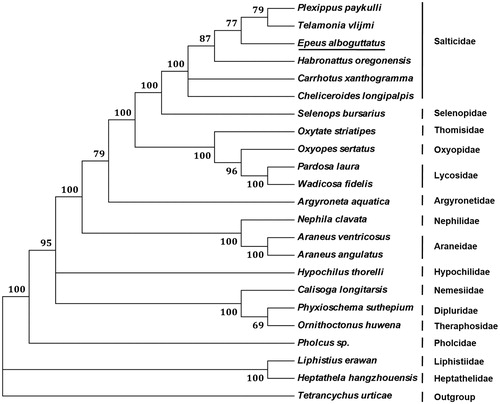
Phylogenetic analysis of E. alboguttatus with 21 other representative spiders using neighbour-joining method based on concatenated amino acid sequences of 13 PCGs showed that E. alboguttatus is nested with Salticidae (), supporting classification based on morphological features. Within the Salticidae clade, E. alboguttatus is recovered as sister to other five jumping spiders.
Figure 1. Phylogenetic tree showing the relationship between E. alboguttatus and 21 other spiders based on neighbour-joining method. Tetranychus urticae was used as an outgroup. GeneBank accession numbers used in the study are the following: Plexippus paykulli (KM114572), Telamonia vlijmi (NC_024287), Habronattus oregonensis (AY571145), Carrhotus xanthogramma (KP402247), Cheliceroides longipalpis (MH891570), Selenops bursarius (KM114573), Oxytate striatipes (KM507783), Oxyopes sertatus (KM272950), Pardosa laura (KM272948), Wadicosa fidelis (NC_026123), Argyroneta aquatica (KJ907736), Nephila clavata (NC_008063), Araneus ventricosus (KM588668), Araneus angulatus (KU365988), Hypochilus thorelli (NC_010777), Calisoga longitarsis (NC_010780), Phyxioschema suthepium (NC_020322), Ornithoctonus huwena (NC_005925), Pholcus sp. (KJ782458), Liphistius erawan (NC_020323), Heptathela hangzhouensis (AY309258), and Tetranychus urticae (EU345430). Spider determined in this study was underlined.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Maddison WP, Bodner MR, Needham KM. 2008. Salticid spider phylogeny revisited, with the discovery of a large Australasian clade (Araneae: Salticidae). Zootaxa. 1893:49–64.
- Maddison WP, Hedin MC. 2003. Jumping spider phylogeny (Araneae: Salticidae). Invert Systematics. 17:529–549.
- Pan WJ, Fang HY, Zhang P, Pan HC. 2014. The complete mitochondrial genome of pantropical jumping spider Plexippus paykulli (Araneae: Salticidae). Mitochondrial DNA A DNA Mapp Seq Ana. 27:1–2.
- Zhu XL, Zhang ZS. 2017. The complete mitochondrial genome of Agelena silvatica (Araneae: Agelenidae). Mitochondrial DNA B Resour. 2:58–59.
