Abstract
Deutzianthus tonkinensis (Euphorbiaceae) is an endangered but commercially popular oil-producing tree, whose populations in China are now fragmented, with the total number of individuals decreasing steadily. Here, we characterized the chloroplast (cp) genome of D. tonkinensis using the genome skimming approach. The whole cp genome is 163,481 bp and comprises 134 genes, including eight unique rRNAs, 36 tRNAs, 84 protein-coding genes, and two pseudo-genes (ycf15 and rps16). The overall guanine-cytosine content of D. tonkinensis cp genome is 35.7%. The phylogenetic analysis suggested that D. tonkinensis is closely related to the Vernicia genus. This study will be useful for future studies on endangered oil-producing tree species, and provide a basis for conservation of this important resource plant.
Main text
Deutzianthus tonkinensis Gagnep., an endangered, key species listed in the National secondary protected plants list, mainly occurs in the tropical areas of south China (Wu Citation2007). It has high economic value for the oil industry and landscaping (Guo et al. Citation2009). However, wild populations of D. tonkinensis have been decreasing because of the habitat destruction. Thus, effective conservation methods are necessary for this endangered tree species. There are very few studies focussing on this oil-producing tree, except some morphological studies (van Welzen and Stuppy Citation1999; Ren Citation2008). Therefore, documentation of the complete chloroplast (cp) genome is necessary to develop a conservation strategy for this endangered tree. In this study, we sequenced, assembled, and annotated the complete cp genome of D. tonkinensis using the Illumina pair-end sequencing and genome skimming.
Fresh leaves of D. tonkinensis were collected from Nonggang natural reserve (106.7375 E, 22.2472 N), Guangxi, China. Voucher specimens were deposited in the Herbarium of Xinjiang University, Urumqi, Xinjiang (Accession Number. Shili033). Total genomic DNA was extracted using the traditional cetyl trimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1987). Sequencing was conducted on the Illumina Hiseq Platform (Illumina, San Diego, CA). In total, we obtained 10.0 GB raw data and approximately 20 million high-quality clean reads, with trimmed and quality filtered adapters, were collected using genome skimming sequencing. The cp genome was assembled using the Velvet software (Zerbino and Birney Citation2008). Annotation analysis was performed with Plastome Annotator (Plann) (Huang and Cronk Citation2015). Then, we analysed and corrected the annotations with Geneious (Kearse et al. Citation2012). Next, we generated a physical map of the genome using Organellar Genome DRAW (OGDRAW) (Lohse et al. Citation2013). A Bayesian tree with posterior probabilities (PP) was reconstructed with MrBayes version 3.1.2 (Ronquist et al. Citation2012) using plastid genomes of 19 species of nine angiosperm families. Finally, the complete genome sequence and gene annotations were submitted to GenBank under the accession numbers of MH933861 for D. tonkinensis.
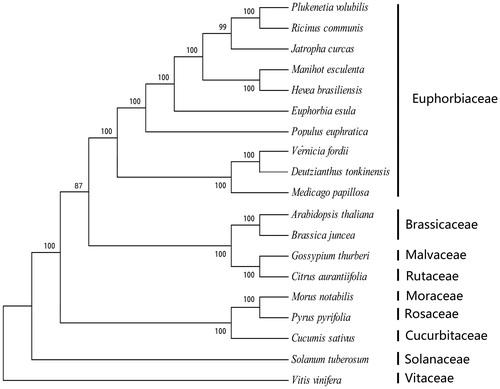
The cp genome of D. tonkinensis is a typical quadripartite structure with a length of 163,481 bp, which contains inverted repeats (IRs) of 26,776 bp separated by a large single copy (LSC) of 91,453 bp and a small single copy (SSC) of 18,476 bp. The cpDNA contains 134 genes, comprising 84 protein coding genes, 36 tRNA genes, eight rRNA genes, and two pseudo-genes (ycf15 and rps16). Among these annotated genes, 11 genes contain one intron (rps12, rpl16, petD, petB, trnV-UAC, trnL-UAA, rpoC1, atpF, trnK-UUU, ndhA, and trnV-UAC), whereas six genes (ycf3, trnA-UGC, trnI-GAU, rpl2, ndhB, and clpP) contain two introns. The overall guanine-cytosine (GC) content of the plastome is 35.7%. Additionally, phylogenetic analysis of 19 plastid genomes where Vitis vinifera was used as an out-group showed that the cp genome of D. tonkinensis is closely related to the Vernicia genus (). Such genetic data can be important to formulate potential new conservation and management strategies for this economically important oil-producing tree.
Figure 1. The phylogenetic tree based on the 19 complete chloroplast genome sequences. Accession numbers: Deutzianthus tonkinensis (MH933861), Manihot esculenta (EU117376), Jatropha curcas (FJ695500), Euphorbia esula (NC033910), Vernicia fordii (KY628420), Plukenetia volubilis (MF062253), Hevea brasiliensis (HQ285842), Medicago papillosa (NC027154), Cucumis sativus (DQ865976), Arabidopsis thaliana (AP000423), Brassica juncea (NC028272), Gossypium thurberi (GU907100), Citrus aurantiifolia (KJ865401), Medicago papillosa (NC027154), Morus notabilis (NC_027110.1), Pyrus pyrifolia (NC015996), Populus euphratica (NC024747), Solanum tuberosum (DQ386163), Vitis vinifera (DQ424856).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guo LF, He JX, Wang XG, Lin CR, HE CX. 2009. Research progress on exploitation of main oil plants of Euphorbiaceae. China Olsand Fats. 34:57–61.
- Huang DI, Cronk QCB. 2015. Plann: A command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:w575–w581.
- Ren MX. 2008. Stamen fusion in plants: diversity, adaptive significance, and taxonomic implications. J Syst Evol. 46:452–466.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- van Welzen PC, Stuppy W. 1999. Phylogenetic considerations of Euphorbiaceae tribe Aleuritideae. Missouri Botanical Garden. 86:894–903.
- Wu ZY. 2007. Flora of China, Euphorbiaceae. Beijing: Science Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
