Abstract
The complete mitochondrial genome (mitogenome) sequence of Sphenomorphus incognitus was determined by shotgun sequencing. The total length of mitogenome is 17 417 bp, and contains 13 protein-coding genes, 22 tRNA genes, two ribosome RNA genes and two non-coding regions (the control region and the putative L-strand replication origin). Most of the genes of S. incognitus were distributed on the H-strand, except for the ND6 subunit gene and eight tRNA genes which were encoded on the L-strand. The phylogenetic tree of S. incognitus and 8 other closely species was built. The DNA data presented here will be useful to study the evolutionary relationships and genetic diversity of S. incognitus.
The genus Sphenomorphus is one of the most diverse groups of lizards with more than 120 known species (Nguyen et al. Citation2011; Roy et al. Citation2013; Uetz et al. Citation2018). None complete mitochondrial genome was reported about this genus. Sphenomorphus incognitus was described by Thompson (1912) from Hengchun, Taiwan, China. This species is currently distributed in northeast of Vietnam and southeast of China, including Taiwan, Hong Kong Hainan, Fujian, Guangxi, Yunnan, Hubei and Anhui (Zhao et al. Citation1999; Nguyen et al. Citation2012; Tang and Huang Citation2014). In this article, we determined and described the mitogenome of S. incognitus in order to obtain basic genetic information about this species.
The specimen of S. incognitus (Voucher number: HUM2012001) was collected from Lingnan Nature Reserve (29°25′50″ N, 118°11′24″ E, Huangshan, Anhui, China) as a new record to Anhui Province and was preserved and deposited in the Museum of Huangshan University (Tang and Huang Citation2014). The complete mitogenome of S. incognitus (Genbank accession number MH329292) was sequenced to be 17, 417 bp which consisted of 13 typical vertebrate protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one D-loop, which is similar to the typical mtDNA of other vertebrates (Boore Citation1999; Sorenson et al. Citation1999).
The positions of RNA genes were predicted by the MITOS (Bernt et al. Citation2012), and the locations of protein-coding genes were identified by comparing with the homologous genes of other closely related species. Most of the S. incognitus mitochondrial genes are encoded on the H-strand except for the ND6 gene and eight tRNA genes, which are encoded on the L-strand. Among the mitochondrial protein-coding genes, the ATP8 was the shortest, while the ND5 was the longest. All protein-coding genes initiate with ATG as start codon, except for COI gene, which began with GTG. Nine genes (ND1, ND2, COI, ATPase 8, ATPase 6, ND4L, ND5, ND6, and Cytb) end with complete stop codons (TAA, AGA, and AGG), and the other four genes end with T as the incomplete stop codons, which were presumably completed as TAA by post transcriptional polyadenylation (Anderson et al. Citation1981). The 22 tRNA genes range in size from 64 to 75 bp. The 12S rRNA (941 bp) and 16S rRNA (1 537 bp), are located between the tRNA-Phe and tRNA-Lue genes and separated by the tRNA-Val gene. The D-loop of the S. incognitus mitogenome in size is 2018 bp and was located between the tRNA-Pro and tRNA-Phe genes. The overall base composition of the entire genome was as follows: A (30.6%), T (25.5%), C (28.8%) and G (15.1%), which the percentage of A + T (56.1%) reflected a typical sequence feature of the vertebrate mitogenome.
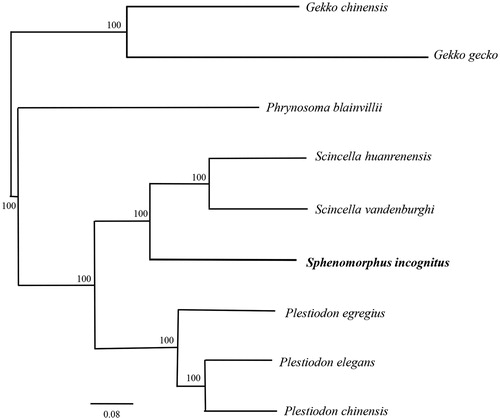
In order to validate the new determined sequences, whole mitochondrial genome sequences of the S. incognitus determined in this study and together with other 8 closely related species from GeneBank to perform phylogenetic analysis. These species were as follows: Gekko chinensis, G. gecko, Phrynosoma blainvillii, Scincella huanrenensis, S. vandenburghi, Sphenomorphus incognitus, Plestiodon egregius, P. elegans, P. chinensis. A maximum likelihood (ML) tree was constructed based on the dataset by online tool RAxML (Stamatakis et al. Citation2008) (). The phylogenetic analysis result was consistent with the previous research. It indicated that our new determined mitogenome sequences could meet the demands and explain some evolution issues.
Figure 1. A maximum-likelihood (ML) tree of the Sphenomorphus incognitus in this study and other eight closely related species was constructed based on the dataset of the whole mitochondrial genome by online tool RAxML. The numbers above the branch meant bootstrap value. Bold black branches highlighted the study species and corresponding phylogenetic classification. The analysed species and corresponding NCBI accession number as follows: Gekko chinensis (KP666135), G. gecko (AY282753), Phrynosoma blainvillii (MG387969), Scincella huanrenensis (KU507306), S. vandenburghi (KU646826), Sphenomorphus incognitus (MH329292), Plestiodon egregius (AB016606), P. elegans (KJ643142), P. chinensis (KT279358).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2012. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Nguyen TQ, Schmitz A, Nguyen TT, Orlov NL, Böhme W, Ziegler T. 2011. A review of the genus Sphenormorphus Fitzinger, 1843 (Squamata: Sauria: Scincidae) in Vietnam, with description of a new species from northern Vietnam and Hainan Island, southern China and the first record of S. mimicus Taylor, 1962 from Vietnam. J Herpetol. 45:145–154.
- Nguyen TQ, Tran TT, Nguyen TT, Ziegler T, Böhme W. 2012. First record of Sphenomorphus incognitus (Thompson, 1912) (Squamata: Scincidae) from Vietnam with some notes on natural history. Asian Herpetol Res. 3:147–150.
- Roy AD, Das I, Bauer AM, Tron RKL, Karanth P. 2013. Lizard wear shades. A spectacled Sphenomorphus (Squamata: Scincidae), from the sacred forests of Mawphlang, Meghalaya, Northe-ast India. Zootaxa. 3071:257–276.
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. 1999. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 12:105–114.
- Stamatakis A, Hoover P, Rougemont J. 2008. A Rapid Bootstrap Algorithm for the RAxML Web-Servers. Syst Biol. 75:758–771.
- Tang XS, Huang S. 2014. Sphenomorphus incognitus firstly found in Anhui Province, China. Chin J Zool. 49:609–612.
- Uetz P, Freed P, Hošek J. 2018. The Reptile Database [accessed 2018 Sep 22]. http://reptile–database.org.
- Zhao EM, Zhao KT, Zhou KY, et al. 1999. Fauna Sinica. Reptilia, Vol.2. Beijing: Science Press, 336–340.
