Abstract
Poaching of wildlife for local consumption and commercial purposes is a major threat to the loss of the world's biodiversity which has even resulted in the extirpation of the numerous species at local or regional scale. Law enforcement agencies have been working in the implementation of the Wildlife Protection Act to combat illegal trade of wildlife for parts and products all across the world. Nevertheless, several times confiscated materials have been altered and fabricated so much that their morphological identification often got difficult. In the last few decades, DNA technology has revolutionized species identification in forensic investigations and has shown its power in the firm identification of those fabricated materials. We received a confiscated material, appeared to be chopped raw meat with no intact morphological identity. The accused was caught by the officials of the State Forest Department after he put pictures on social media depicting his involvement in cooking and eating of animal meat, suspected to be of wildlife origin. On homology search, we found a 99% identity of the confiscated material with Asian Palm Civet (Paradoxurus hermaphroditus) and neighbour joining trees based on genetic distances clustered the query sequence with the Asian Palm Civet with 100% bootstrap support. The present study exhibits hope for ascertaining species identification from the processed meat in reliable assessment for wildlife forensics. This case study extends, the role of the authenticated references as available on the public database, highlighting the application of DNA forensics in identifying species even from the phenotypically altered materials.
Introduction
Poaching of wild animals for meat, skin, bone, and other body parts is one of the most ancient cultures on the way of human civilization and evolution (Robert and John Citation1998). This regular practices with the inclusion of commercial greed of mankind has caused the loss of biodiversity and brought several species extinct like Javan Rhino (Streicher et al. Citation2010), Western Black Rhino (Lagrot et al. Citation2007), Passenger pigeon (Hume Citation2015), or near extinct like Pink-Headed Duck (Birdlife International Citation2016), Hangul ). Although, all wild animals in India are protected by the Wildlife (Protection) Act, 1972. However, illegal poaching and trade for wildlife parts and products are still silently on, and several animals are being poached every year (Thakur Citation2014). Examination in forensics based on morphological and immunological clues has limited power in ascertaining species and individual identity using conventional methods. In wildlife forensics, species identification from parts and products which often got altered and processed several folds, become challenging using conventional methods even for a trained and skilled biologist. Often, specimens received for identification are either too old and degraded (Alacs et al. Citation2010) or altered into pieces (Thakur et al. Citation2017) that ascertaining species identity becomes very difficult. Therefore, DNA-based identifications become a useful tool for the investigation of confiscated wildlife samples with altered morphological identity (Verma and Singh Citation2002; Gupta et al. Citation2011). Mitochondrial genes like CoI (Hebert et al. Citation2003; Khedkar et al. Citation2016) and CytB (Jun et al. Citation2011; Kumar et al. Citation2016a; Muangkram et al. Citation2018), 16S rRNA (Guha and Kashyap Citation2006; Kumar et al. Citation2016b), and 12S rRNA (Mukesh et al. Citation2013; Thakur et al. Citation2017) have been used for catering the need of species identification from the confiscated materials and have been proven reliable for species identification from a variety of samples with compromised morphological identity. The present study was aimed to identify the species of origin from an altered confiscated material to submit a technical advisory to prosecute legal trials in the court of law and for the implementation of the Wildlife (Protection) Act, 1972 of India.
Case presentation
A person from Palta (North 24 Parganas District) of West Bengal State, India was suspected to poach an animal of wild origin as he was caught by the forest department after he uploaded few photos of himself in the act of killing and preparing to cook a wild animal on social media. The State Forest Department arrested him just after immediate hype on social media under the violation of Wildlife (Protection) Act, 1972. Divisional Forest Officer of North 24 Parganas, West Bengal, confiscated the meat, presumed of wildlife origin and sent it to Zoological Survey of India, Kolkata for species identification via letter no. 1602(3)/15-8, dated 23 November 2017.
Materials and methods
We took a subsample of meat, chopped into small pieces, coded as ‘ZSI/FI/2017/11’ and washed overnight with 1X sterile Phosphate Buffer Saline (PBS). Isolation of genomic DNA was carried out through Phenol-Chloroform-Isoamyl alcohol (PCI) method (Sambrook Citation1982; Ausubel et al. Citation1997). Universal primers of Cytochrome B (Forward primer Mcb398 5′-TACCATGAGGACAAATATCATTCTG-3′ and reverse primer Mcb869 5′-CCTCCTAGTTTGTTAGGGATTG ATCG-3′; Verma and Singh Citation2002) and 12S rRNA genes (forward primer 5′-CAAACTGGGATTAGATACCCCACTAT-3′ and Reverse primer 5′-GAGGGTGACGGGCGGTGTGT-3′; Kocher et al. Citation1989) were used to amplify these genes for species-level identification.
PCR products were cleaned up using Exo-SAP treatment, and Cycle sequencing was carried out independently for forward and reverse primer using Big Dye terminator cycle sequencing kit v3.1 (Applied Biosystems, Foster City, CA). Sequencing was performed on a Genetic analyser 3730 (Applied Biosystems, Foster City, CA). The sequence quality was checked manually by nucleotide and validated using Sequencher v4.7 (www.genecode.com). Homologous sequences were downloaded from NCBI/BLAST search, and multiple sequence alignment for all the downloaded sequences was carried out with MUSCLE alignment tool in MEGA6 (Tamura et al. Citation2013). To confirm the species identity, we undertook phylogenetic re-construction of the confiscated material with the downloaded sequences. The best fit model for constructing phylogeny was determined based on the lowest BIC value, and the neighbour joining (NJ) trees were constructed in MEGA6 following Tamura-Nei model with a gamma distribution for both the genes.
Results
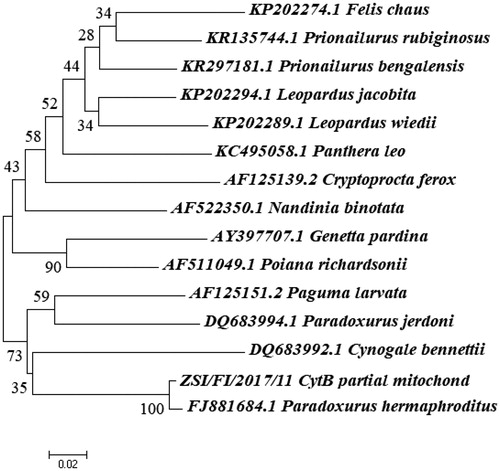
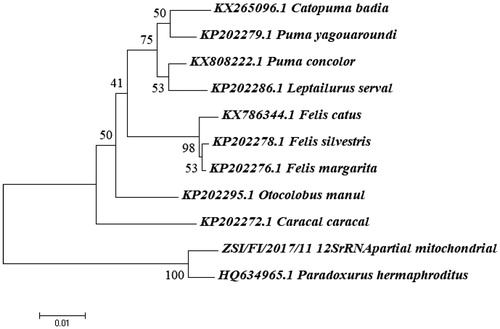
DNA from the confiscated meat sample successfully isolated using PCI method and visualized on 1% agarose gel. PCR amplified a fragment of both CytB and 12S rRNA genes visualized (on 2% agarose gel) as separate single DNA bands between 250 bp and 500 bp band of the 1 kb ladder. On BLAST search for both the genes, the query sequences showed to have 99% identity with the sequences of Asian Palm Civet (Paradoxurus hermaphroditus). The genetic distance matrix for both the genes revealed that the suspected confiscated material have closest genetic similarity (Nei's DA 0.011 for CytB gene) and (Nei's DA 0.012 for 12S rRNA) with the known sequences of Asian Palm Civet, Paradoxurus hermaphroditus and NJ trees demonstrated that query sequences clustered tightly with the known sequences of Asian Palm Civet (Paradoxurus hermaphroditus) with 100% bootstrap support ( and ). Hence, this got confirmed by the collective evidences like homology search on NCBI/Genbank database, genetic distances and phylogenetic tree analysis that the confiscated material is of Asian Palm Civet Origin Paradoxurus hermaphroditus which is scheduled species (Schedule II) and protected under Wildlife (Protection) Act, 1972 and widely distributed in the State of West Bengal. The case report on species identification was submitted to the authority of the Forest Department of West Bengal. The analsed sample in the form of isolated DNA and the tissue stored in alcohol has been archived in the reference repository of confiscated material at Centre for DNA Taxonomy of ZSI, Kolkata. The resulted sequences of Cytochrome b and 12Sr RNA gene of Asian Palm Civet were submitted to NCBI/GenBank database with the Accession No. MH423777 and MH423735.
Discussion
Although, several earlier studies have shown the applicability of barcoding genes for rapid and reliable forensic identification (Verma and Singh Citation2002; Mukesh et al. Citation2013), the present study has shown an extended validation of the technique in identifying species from raw meat which had lost the morphological identity before processing the sample in laboratory. Hence, the current study generated new hope by showing that documenting species identification is possible even for raw meat for reliable assessment of wildlife forensic investigations.
Acknowledgements
Authors thank the forest officials, 24 North Parganas, West Bengal for providing the case property for species identification.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alacs EA, Georges A, FitzSimmons NN, Robertson J. 2010. DNA detective: a review of molecular approaches to wildlife forensics. Forensic Sci Med Pathol. 6:180–194.
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1997. Current protocols in molecular biology. Hoboken (NJ): John Wiley & Sons, Inc.
- Birdlife International. 2016. Rhodonessa caryophyllacea. The IUCN Red List of Threatened Species 2016. Downloaded on 06 June 2018.
- Guha S, Kashyap VK. 2006. Molecular identification of lizard by RAPD & FINS of mitochondrial 16s rRNA gene. Leg Med (Tokyo). 8:5–10.
- Gupta SK, Thangaraj K, Singh L. 2011. Identification of the source of ivory idol by DNA analysis. J Forensic Sci. 56:1343–1345.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Biol Sci. 270:313–321.
- Hume JP. 2015. Large-scale live capture of Passenger Pigeons Ectopistes migratorius for sporting purposes: overlooked illustrated documentation. Bull BritOrnithologists Club. 135:174–184.
- Jun J, Han SH, Jeong TJ, Park HC, Lee B, Kwak M. 2011. Wildlife forensics using mitochondrial DNA sequences: species identification based on hairs collected in the field and confiscated tanned Felidae leathers. Genes Genomics. 33:721–726.
- Khedkar GD, Abhayankar SB, Nalage D, Ahmed SN, Khedkar CD. 2016. DNA barcode based wildlife forensics for resolving the origin of claw samples using a novel primer cocktail. Mito DNA Part A. 27:3932–3935.
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC. 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers, Evolution. Proc Natl Acad Sci USA. 86:6196–6200.
- Kumar VP, Rajpoot A, Mukesh, Kumar D, Goyal SP. 2016a. Genetic based species identification and tracking of the geographic origin of a fully tanned animal skin in wildlife forensics. Forensic Res Criminol Int J. 2:00058.
- Kumar VP, Rajpoot A, Mukesh, Shukla M, Kumar D, Goyal SP. 2016b. Illegal trade of Indian Pangolin (Manis crassicaudata): genetic study from scales based on mitochondrial genes. Egy J Foren Sci. 6:524–533.
- Lagrot I, Lagrot JF, Bour P. 2007. Probable extinction of the western black rhino, Diceros bicornis longipes: 2006 survey in northern Cameroon. Pachyderm. 43:19–28.
- Muangkram Y, Wajjwalku W, Amano A, Sukmak M. 2018. The novel primers for mammal species identification-based mitochondrial cytochrome b sequence: implication for reserved wild animals in Thailand and endangered mammal species in Southeast Asia. Mito DNA Part A. 29:62–72.
- Mukesh SSK, Shukla M, Sharma LK, Mohan N, Goyal SP, Sathyakumar S. 2013. Identification of galliformes through forensically informative nucleotide sequencing (FINS) and its implication in wildlife forensics. J Forensic Res. 4:195.
- Robert MM, John Jr FB. 1998. Illegal harvest of renewable natural resources in North America: toward a typology of the motivations for poaching. Soc Natural Res. 11:9–24.
- Sambrook J, Fritsch EF, Maniatis T. 1982. Molecular cloning: a laboratory manual. New York (NY): Cold Spring Harbor Laboratory Press.
- Streicher U, Newcomer E, McCarty D, Brook S, Hai Bach Thanh. 2010. Examination of a Dead Javan Rhinoceros, Cat Tien National Park Vietnam. WWF Vietnam Programme, Hanoi, Vietnam.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Thakur M. 2014. Role of DNA Forensics in curbing illegal wildlife trade. WWF Newsletter PANDA, Illegal Wildlife Trade in India: Special Issue. p. 11–12.
- Thakur M, Javed R, Kumar VP, Shukla M, Singh N, Maheshwari A, Mohan N, Wu DD, Zhang YP. 2017. DNA forensics in combating food frauds: a study from China in identifying canned meat labelled as deer origin. Curr Sci. 112:2449–2452.
- Verma SK, Singh L. 2002. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol Ecol Notes. 3:28–31.