Abstract
We report the first complete mitochondrial genome of Callogorgia. Callogorgia cf. gracilis isolate DFH32_518B was collected by a remotely operated vehicle at 98 m on McGrail Bank (27.9840725°N, 92.604242°W). The complete mitogenome is 18,937 bp (27.8% A, 18.3% C, 19.8% G, and 34.1% T) and has the ancestral octocoral gene order for its 14 protein-coding genes, two rRNA genes, and one tRNA gene. It is sister to and ∼96.6% similar (uncorrected) to Narella hawaiiensis, the only other complete mitogenome reported for Primnoidae. The cox1 + igr1 + mtMutS region differs by two base pairs (0.12%) from the only reported C. gracilis haplotype.
Callogorgia consists of 27 valid species (Cairns et al. Citation2018), four of which occur in the northwestern Gulf of Mexico (Cairns and Bayer Citation2002; Etnoyer and Cairns Citation2017): C. americana, C. delta, C. gracilis, and C. linguimaris. C. gracilis is the only one reported shallower than 180 m (Quattrini et al. Citation2013; Etnoyer and Cairns Citation2017) and is distinguishable by its stiff, straight main stem, cylindrical (vs. clavate) polyps, number of sclerites, and unique mitochondrial molecular barcode (cox1+igr1+mtMutS) (Cairns and Bayer Citation2002; Quattrini et al. Citation2013). The collected specimen is morphologically consistent with C. gracilis but differs by two base pairs (0.12%) from the single cox1+igr1+mtMutS haplotype reported in Quattrini et al. (Citation2013) for C. gracilis (DOI: 10.6084/m9.figshare.6998459). Because additional specimens are required to delineate species within the C. gracilis complex (see Cairns and Bayer Citation2002), we assigned this specimen to C. cf. gracilis.
The specimen was collected by remotely operated vehicle (SubAtlantic Mohawk 18 operated by University of North Carolina – Wilmington, Undersea Vehicles Program) at 98 m on McGrail Bank (27.9840725°N, 92.604242°W) on 24 September 2017. DNA was extracted with GeneJET Genomic DNA Purification Kit (ThermoFisher Scientific Waltham, MA) per manufacture’s protocol and submitted to Biopolymers Facility at Harvard Medical School for library preparation and next-generation sequencing (NextSeq 500). Trimmed reads (Trimmomatic-0.32, Bolger et al. Citation2014) were assembled de novo with SPAdes (Bankevich et al. Citation2012) on the University of New Hampshire Bioinformatics Core facility ron server. After circularizing and editing overlapping ends of the SPAdes contig in Geneious R10.2.6 (Kearse et al. Citation2012), trimmed reads (BBDuk v. 37.25) were mapped to the resulting reference sequence to generate a consensus sequence. Genes were annotated by manually adjusting Narella hawaiiensis (NC026192) annotations mapped to the consensus sequence. The complete mitogenome was aligned with default MUSCLE (Edgar Citation2004) parameters to eight representative species with the octocoral ancestral mitochondrial gene order and complete mitogenomes available in GenBank: all Calcaxonia species, one randomly selected representative from each Alcyonacea subclass, and all Helioporacea and Pennatulacea species. The latter two taxa generally are sister to the clade containing Primnoidae (Brockman and McFadden Citation2012; Poliseno et al. Citation2017). A maximum-likelihood, phylogenetic tree was constructed in RAxML 8.2.11 (Stamatakis Citation2014) (). Extended methods and cox1+igr1+mtMutS alignment are available at figshare (DOI: 10.6084/m9.figshare.6998459).
Figure 1. Maximum-likelihood, phylogenetic tree of the complete mitogenomes of Callogorgia cf. gracilis (this study) and eight representative octocorals (taxonomic position and GenBank accession numbers in tip labels). In Geneious R10.2.6, complete mitogenomes were aligned with default MUSCLE parameters; the resulting alignment was used to construct the phylogenetic tree in RAxML 8.2.11 with the following changes to the default settings: bootstrap replicates =1000, algorithm = rapid bootstrapping and search for best-scoring ML tree, outgroup = NC023344, nucleotide model = GTR CAT I. Bootstrap values are reported at the nodes. See figshare (DOI: 10.6084/m9.figshare.6998459) for extended methods.
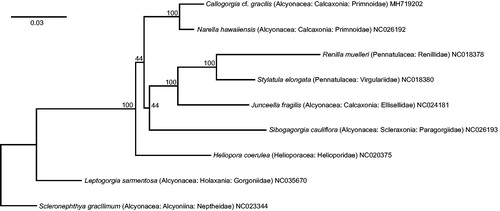
The complete mitogenome is 18,937 bp (27.8% A, 18.3% C, 19.8% G, and 34.1% T), has the ancestral octocoral gene order, and has one tRNA, two rRNA, and 14 protein-coding genes. Despite the utility of gene-sequence data in resolving coral phylogenies (e.g. Soler-Hurtado et al. Citation2017), this mitogenome report is the first for Callogorgia and second for Primnoidae. C. cf. gracilis is sister to and ∼96.6% similar (uncorrected) to the only other Primnoidae, N. hawaiiensis, but the relative position of Primnoidae among octocoral families is not well supported (). The C. cf. gracilis mitogenome was deposited in GenBank (MH719202) and the specimen in the Smithsonian National Museum of National History (USNM1507967).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Brockman SA, McFadden CS. 2012. The mitochondrial genome of Paraminabea aldersladei (Cnidaria: Anthozoa: Octocorallia) supports intramolecular recombination as the primary mechanism of gene rearrangement in octocoral mitochondrial genomes. Genome Biol Evol. 4:994–1006.
- Cairns SD, Bayer FM. 2002. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 2: The genus Callogorgia Gray, 1858. Proceedings of the Biological Society of Washington. 115:840–867.
- Cairns SD, Stone RP, Moon H-W, Lee JH. 2018. Primnoidae (Octocorallia: Calcaxonia) from the Emperor Seamounts, with notes on Callogorgia elegans (Gray, 1870). Pac Sci. 72:125–142.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Etnoyer PJ, Cairns SD. 2017. Deep-sea coral taxa in the US Gulf of Mexico: depth and geographical distribution. https://deepseacoraldata.noaa.gov/. [Last accessed on 6 August 2018].
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Poliseno A, Feregrino C, Sartoretto S, Aurelle D, Wörheide G, McFadden CS, Vargas S. 2017. Comparative mitogenomics, phylogeny and evolutionary history of Leptogorgia (Gorgoniidae). Mol Phylogenet Evol. 115:181–189.
- Quattrini AM, Georgian SE, Byrnes L, Stevens A, Falco R, Cordes EE. 2013. Niche divergence by deep-sea octocorals in the genus Callogorgia across the continental slope of the Gulf of Mexico. Mol Ecol. 22:4123–4140.
- Soler-Hurtado MM, López-González PJ, Machordom A. 2017. Molecular phylogenetic relationships reveal contrasting evolutionary patterns in Gorgoniidae (Octocorallia) in the Eastern Pacific. Mol Phylogenet Evol. 111:219–230.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
