Abstract
Periploca sepium Bunge is not only a traditional medicinal plant, but also a pioneer species for soil and water conservation because of its great environmental adaptability. In this study, its complete chloroplast genome was assembled from the whole genome Illumina sequencing data. The circular genome was 153,513 bp in size, containing a large single copy (LSC) region of 83,818 bp and a small single copy (SSC) region of 18,177 bp, which were separated by a pair of 25,759 bp inverted repeat (IR) regions. It encoded a total of 139 genes, including 85 protein-coding genes, 46 tRNA genes, and eight rRNA genes. The most of gene species occurred as a single copy, while 15 gene species occurred in double copies. The overall A + T content was 61.9%, while the corresponding values of the LSC, SSC, and IR regions were 63.8, 68.1, and 56.6%, respectively. Phylogenetic analysis indicated that P. sepium was relatively close to the species belonging to the subfamily Apocynoideae.
Periploca sepium Bunge is a traditional medicinal plant of the Periplocoideae within the family Apocynaceae, which is widely distributed in the northern temperate regions of China (Li et al. Citation1995). Its dried root bark is used as a Chinese medicinal herb for the treatment of rheumatism, cancer, and cardiac failure (Yin et al. Citation2009; Ding et al. Citation2014). Due to its strong environmental adaptability such as cold and drought tolerance and sand resistance, this species is also used as a pioneer plant for soil and water conservation (Yang et al. Citation2006; An et al. Citation2007). Despite its great medicinal and ecological value, the good knowledge of genetics and genomics for this plant is still lacking. In order to resolve the interspecific relationship in crown clade Apocynaceae, a partial chloroplast genome of P. sepium was reported by Straub et al. (Citation2014). However, the problem of its taxonomical phylogenetical position is still doubtful due to the incomplete genomic information. In the present study, the complete chloroplast genome sequence of P. sepium is reported for contributing to the conservation and sustainable utilization of this species, and also providing significant information for its phylogenetic placement within the family Apocynaceae.
Genomic DNA was isolated from fresh leaves of an individual P. sepium collected from the Qinling Mountain (108°32'24"E, 33°33'48"N; the specimen was deposited at Shaanxi Normal University; accession number: ZJB-2017-163-1). The whole-genome sequencing was conducted on an Illumina Hiseq X Ten platform. The resultant clean reads were then assembled into complete chloroplast genome with the program Velvet (Zerbino and Birney Citation2008), with Asclepias syriaca (GenBank: KF386166.1) as the starting reference. The complete chloroplast genome was annotated using Geneious (Kearse et al. Citation2012) and then submitted to GenBank (accession no. MH752592).
The complete chloroplast genome of P. sepium was 153,513 bp in size, including a large single copy (LSC, 83,818 bp) region, a small single copy (SSC, 18,177 bp) region, and two inverted repeat (IR, 25,759 bp) regions. The circular genome contained 139 genes, including 85 protein-coding genes (81 PCG species), eight rRNA genes (4 rRNA species), and 46 tRNA genes (31 tRNA species). The most of gene species occurred in a single copy, while 15 gene species occurred in double copies, including four rRNA species (4.5S, 5S, 16S, and 23S rRNA), seven tRNA species and four PCG species. The overall A + T content was 61.9%, while the corresponding values of the LSC, SSC, and IR regions were 63.8, 68.1, and 56.6%, respectively.
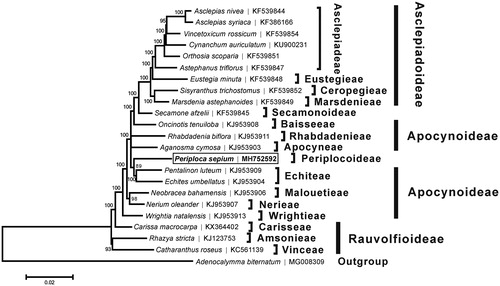
A neighbor-joining tree () was reconstructed based on the complete chloroplast genome sequences of P. sepium and other 21 species from Apocynaceae with the program MEGA6 (Tamura et al. Citation2013). The phylogenetic analysis supported the traditional taxonomy of the family Apocynaceae at the genus level, but failed so at the subfamily level. However, this result was consistent with the phylogenic study suggested by Straub et al. (Citation2014), P. sepium was found to be relatively closely related to two species (Pentalinon luteum and Echites umbellatus) belonging to the subfamily Apocynoideae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- An Y, Liang Z, Han R, Liu G. 2007. Effects of soil drought on seedling growth and water metabolism of three common shrubs in Loess Plateau, Northwest China. Front Forest China. 2:410–416.
- Ding FF, Zhang XJ, Deng YR. 2014. Inhibition of periplocin on human hepatoma carcinoma and breast carcinoma cells in vitro. Drug Eval Res. 37:30–33.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li PT, Gillbert MG, Stevens WD. 1995. Asclepiadaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol.16. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 189–270.
- Straub SC, Moore MJ, Soltis PS, Soltis DE, Liston A, Livshultz T. 2014. Phylogenetic signal detection from an ancient rapid radiation: effects of noise reduction, long-branch attraction, and model selection in crown clade Apocynaceae. Mol Phylogenet Evol. 80:169–185.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yang CH, Wang YY, Zhou ZF, Zhang GC. 2006. Response of gas exchange parameters of Periploca sepium Bunge to soil water content in loess plateau. Forest Res. 19:231–234.
- Yin ZQ, Wang L, Zhang XQ, Ye WC, Zhang J. 2009. Steroids from the roots of Periploca sepium. Chin Pharm J. 44:968–971.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.