Abstract
Castanea seguinii is a Chinese chestnut, also called Seguin chestnut. It is cultivated for its edible nuts by rural people in subtropical regions and in southwestern China. We determined the complete chloroplast genome sequence for C. seguinii using Illumina sequencing data. The complete chloroplast sequence is 161,010 bp in length and contains a pair of 25,701 bp inverted repeats (IRs), a large single-copy (LSC) region of 90,560 bp, and a small single-copy (SSC) region of 19,048 bp. It contains 132 genes, including 103 protein-coding genes, eight ribosomal RNA genes (four RNA species), 46 transfer RNA genes, and one pseudogene (ycf1). The overall GC content of the whole genome is 36.7%. Phylogenetic analysis based on 20 chloroplast genomes indicates that C. seguinii is closely related to Chinese chestnut (C. mollissima).
Castanea seguinii is a small tree or shrub perennial plant belonging to Fagaceae and an endemic chestnut species of China. The species is widely distributed in the south of the Dabie Mountains found in mixed mesophytic forests, thicket, and orchards from 400 to 2000 m in southwestern China (Mellano et al. Citation2012). C. seguinii and C. mollissima are sympatrically distributed in low-altitude hilly regions of east-central China, and the morphological characteristics of C. seguinii are similar with the C. mollissima (Huang et al. Citation1994). C. seguinii had very small nuts (2–4 g), which is cultivated for its edible nuts but not like as extensively as C. mollissima (Mellano et al. Citation2012). In this study, we report the complete chloroplast genome (cp) of C. seguinii based on Illumina pair-end sequencing data.
The plant material of C. seguinii was sampled from Nanchang, Jiangxi Province, China. The voucher specimen is kept at the Evolutionary Botany Laboratory, College of Life Sciences, Northwest University (Xi’an, Shaanxi, China). Genomic DNA was extracted from leaf tissue using the Plant Genomic DNA kit from Tiangen Biotech (Beijing) Co., Ltd. (Beijing, China). The whole-genome sequencing was conducted with 350 bp pair-end reads on the Illumina Hiseq 4000 platform (Illumina, San Diego, CA) by Novogene, Beijing, China.
The high-quality paired-end reads were assembled with the program MITObim v1.7 (Oslo, Switzerland) (Hahn et al. Citation2013) using the C. mollissima chloroplast genome sequence as a reference (GenBank accession HQ336406) (Jansen et al. Citation2011). Annotation was performed using the online program Dual Organellar Genome Annotator (DOGMA) (Austin, United States) (Wyman et al. Citation2004). We generated a physical map of the chloroplast genome using OGDRAW (Lohse et al. Citation2013). The cp genome sequence was submitted to GenBank (accession number MH998383).
The cp genome of C. seguinii was 161,010 bp in length and contains a pair of inverted repeats (IRa and IRb) regions of 25,701 bp, the large single-copy (LSC) region and small single-copy (SSC) region of 19,048 and 90,560 bp, respectively. The genome contains 132 genes, including 103 are protein-coding genes, 46 are transfer RNA genes, and eight ribosomal RNA genes. Among these genes, 14 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) have one intron, and three genes (clpP, rps12, ycf3) have two introns. Overall GC content was 36.7% and in LSC, SSC, and IR regions were 34.6, 30.8, and 42.7%, respectively.
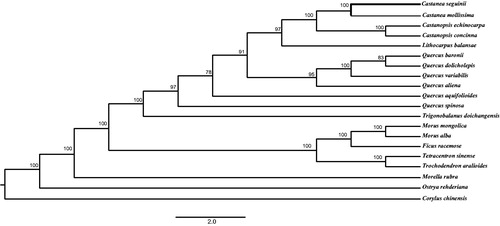
Plastome sequences of C. seguinii plus 11 Fagaceae species and nine other plant species were aligned with MAFFT v7.0.0 (Katoh and Standley Citation2013). The phylogenetic relationship analysis was inferred using the Bayesian inference (BI) method, which was performed using MrBayes v3.2.6 (Stockholm, Sweden) (Ronquist et al. Citation2012). The local bootstrap probability of each branch was calculated by 1000 replications. The phylogenetic tree showed that C. seguinii was most closely related to C. mollissima with 100% bootstrap ().
Figure 1. Bayesian inference (BI) phylogenetic tree based on 20 complete chloroplast genome sequences. Accession numbers: Castanea mollissima HQ336406, Castanopsis echinocarpa NC_023801, Castanopsis concinna NC_033409, Lithocarpus balansae KP299291, Quercus baronii KT963087, Quercus dolicholepis KU240010, Quercus variabilis KU240009, Quercus aliena KU240007, Quercus aquifolioides NC_026913, Quercus spinosa NC_026907, Trigonobalanus doichangensis NC_023959, Morus mongolica KM491711, Morus alba KU355276, Ficus racemose KT368151, Tetracentron sinense KC608752, Trochodendron aralioides KC608753, Morella rubra NC_035006, Ostrya rehderiana NC_028349, and Corylus chinensis KX814336. The number on each node of branch indicates the bootstrap support value. The bond line marks the newly sequenced C. seguinii in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Huang H, Dane F, Norton JD. 1994. Allozyme diversity in Chinese, Seguin and American chestnut (Castanea spp.). Theor Appl Genet. 88:981–985.
- Jansen PK, Saski C, Lee SB, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28:835–847.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molec Biol Evol. 30:772–780.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW - a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Mellano MG, Beccaro GL, Donno D, Marinoni DT, Boccacci P, Canterino S, Cerutti AK, Bounous G. 2012. Castanea spp. biodiversity conservation: collection and characterization of the genetic diversity of an endangered species. Genet Resour Crop Evol. 59:1727–1741.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
