Abstract
The Chinese zokor, Eospalax fontanierii, is a group of common subterranean solitary rodents and endemic in China. The complete mitogenome sequence of E. fontanierii was determined based on next-generation sequencing in this study. The genome was 16,369 bp long, containing 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). The overall base composition of the heavy strand is A (33.80%), C (23.85%), T (30.23%), and G (12.11%). The base compositions present highly biased toward A + T nucleotides. The result of phylogenetic analysis showed the four Eospalax species formed a monophyly with the high bootstrap value, and as a sister group of the genus Myospalax. This is the first report of the complete mitochondrial genomes of E. fontanierii, and the mitogenome is potentially important for evolutionary biology, population genetics, and species diagnosis studies of the Mysopalacinae.
The Chinese zokor, Eospalax fontanierii, is a group of common subterranean solitary rodents belonging to subfamily Myospalacinae, family Spalacidae (Norris et al. Citation2004), distributing in North China (Fan and Shi Citation1982).
In this study, we successfully sequenced the first complete mitogenome of E. fontanierii by next-generation sequencing. The sample was collected from Hebei province of China (N 40°49′16.57″, E 115°48′3.87″, altitude 1122 m), in October 2017. Voucher specimens (No. CCX-3) were placed in Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Total genomic DNA was extracted from liver using DNeasy Tissue Kit (QIAGEN) following the manufacturer’s protocols. An Illumina library was generated from genomic DNA and sequenced on an Illumina Hiseq 2500 platform. The complete mitogenome sequence of E. fontanierii was assembled, annotated and analyzed.
The accurate annotated mitochondrial genome sequence of E. fontanierii was submitted to GenBank with accession number MH884481. The mitochondrial genome is a circular double-strand DNA molecule of 16,369 bp, containing 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and one non-coding region. The arrangement of the multiple genes is similar in line with other Myospalacinae species (Liu et al. Citation2011; Su et al. Citation2013; Li et al. Citation2016; Yuan et al. Citation2016). The overall base composition of the heavy strand is 33.80% A, 23.85% C, 30.23% T, and 12.11% G. The base compositions present highly biased toward A + T nucleotides. The heavy strand (H-strand) encodes two rRNA genes, 12 PCGs, and 14 tRNA genes, the nicotinamide adenine dinucleotide dehydrogenase subunit 6 (ND6) gene and eight other tRNA genes are encoded on the L-strand. For 10 of the 13 protein-coding genes, the start codon is ATG. The ND2 gene starts with ATT, the ND3 gene starts with ATA, and the ND5 gene starts with ATT. Eight of the 13 protein-coding genes use TAA as the stop codon. The ND1, ND2, ND6, and Cytochrome oxidase subunit 3 (COX3) were stopped with TAG, and the ND4 was stopped with AGA. The 12S and 16S ribosomal RNA genes are 944 and 1475 bp long, respectively. The tRNA genes vary from 60 to 75 bp in length and employ the anticodons typical of vertebrate mt-tRNAs. Twenty tRNAs had a typical secondary structure (cloverleaf structure) except the tRNA-Ser and tRNA-Lys, whose complete dihydrouridine arms were lacking. Mismatched G-U pairs, C-U pairs, A-A pairs, and U-U pairs were found in the 22 tRNA secondary structures of E. fontanierii mitogenome. In our study, the noncoding control region of the E. fontanierii mtDNA is 949 bp long.
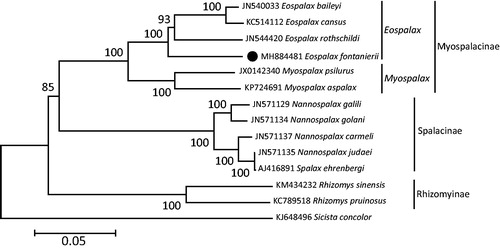
Phylogenetic relationships were inferred from available mitogenomes of 13 species of Spalacidae using Maximum-Likelihood (ML) method with 13 PCGs and two rRNAs. The results of phylogenetic analysis displayed that Myospalacinae contained two monophylies of Myospalax and Eospalax, and Myospalacinae was a sister group with Spalacinae and Rhizomyinae (). This mitogenome sequence of E. fontanierii would help in evolutionary biology, population genetics, and species diagnosis studies of the Mysopalacinae.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of Eospalax fontanierii and the other 12 species of Spalacidae using Sicista concolor as an outgroup. The number around each node indicates the ML bootstrap support values. All 14 species’ accession numbers are listed as below: E. fontanierii (MH884481), E. baileyi (JN540033), E. cansus (KC514112), E. rothschildi (JN544420), Myospalax aspalax (KP724691), M. psilurus (JX014234), Spalax carmeli (JN571137), Nannospalax galili (JN571129), N. golani (JN571134), N. judaei (JN571135), N. ehrenbergi (AJ416891), Rhizomys pruinosus (KC789518), R. sinensis (KM434232), and S. concolor (KJ648496).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Fan N, Shi Y. 1982. A revision of the zokors of subgenus Eospalax. Acta Zool Sin. 2:180–199 (Chinese).
- Liu ZJ, Li YW, Shi FL, Lu JQ, Li M, Wang ZL. 2011. Mitochondrial genome of Plateau zokor Myospalax baileyi. Mitochondrial DNA. 22:174–175.
- Li Y, Lu J, Wang Z. 2016. Complete mitochondrial genome of manchurian zokor (Myospalax psilurus). Mitochondrial DNA. 27:1461–1462.
- Norris RW, Zhou K, Zhou C, Yang G, William KC, Honeycutt RL. 2004. The phylogenetic position of the zokors (myospalacinae) and comments on the families of muroids (rodentia). Mol Phylogenet Evol. 31:972–978.
- Su J, Wang J, Hua L, Gleeson D, Ji W. 2013. Complete mitochondrial genome of the Gansu zokor, Eospalax cansus (rodentia, spalacidae). Mitochondrial DNA. 24:651–653.
- Yuan S, Lu Z, Wu X, Fu H, Bao D, Malqin H, Yang S. 2016. Complete mitochondrial genome of Myospalax aspalax (rodentia, spalacidae). Mitochondrial DNA. 27:4250–4251.
