Abstract
Hypostomus francisci, commonly known as “cascudo”, is a fish of ecological importance found in the upper São Francisco River, Minas Gerais, Brazil. Here we present the complete mitogenome (mtDNA) of H. francisci. The whole molecule is 16,541 bp long and contains 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and one control region (D-loop). All the PCGs in the H. francisci mtDNA use the standard ATG start codon, except for Cox1, which utilizes GTG. Seven of the 13 PCGs contain TAA stop codons, three contain a TAG, and three contain the incomplete stop codons TA − or T−.
Material, results, and discussion
Hypostominae is the richest subfamily within Loricariidae, with 477 valid species (Eschmeyer et al. Citation2017). Hypostomus (Lacépède, 1803) is the most diverse genus, currently comprising about 135 species (Isbrücker Citation2001; Armbruster Citation2004; Dias and Zawadzki Citation2018). H. francisci is the most common Hypostomus species in the basin São Francisco (Lütken Citation1874; Alves and Pompeu Citation2010). The taxonomy of Hypostomus from the São Francisco River is still poorly understood (Ramos et al. Citation2017).
DNA was extracted from the muscle tissue of one H. francisci specimen collected in the Abaeté River, a tributary of the São Francisco River, Minas Gerais, Brazil (UTM 23K 0450882 7997412). The voucher (NUP 20522) was deposited in the collection of the Museum of Nupelia, State University of Maringá, Paraná, Brazil. The genomic library was constructed and sequenced using a paired-end 150 bp strategy in a MiSeq platform (Illumina®). The assembly was generated using the software Mira 4.0 (Chevreux et al. Citation1999) and Geneious R7.1.3 (Drummond et al. Citation2009).
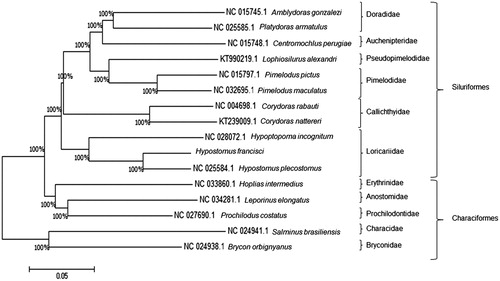
The full mtDNA was 16,541 bp long, resolved with 20× of coverage (GenBank accession number MK026008). The GC content was 41.28%, and individual base frequencies were as follow: 31.42% of A, 27.30% of T, 26.37% of C, and14.91% of G. Annotation was performed in the MitoFish webserver (Iwasaki et al. Citation2013), and the Mitos Web Server (Bernt et al. Citation2013) was used for verification of the start and stop codons. The mitogenome presented two rRNA, 22 tRNA, 13 protein-coding genes (PCGs), and one control region. Genes arrangement was similar to that of a typical vertebrate mitogenome. The gene Cox1 contained a GTG start codon, while all the other PCGs displayed the usual ATG start codon. Seven (ND1, COI, ATP8, ATP6, ND4L, ND5, and CytB) of the 13 PCGs contained a TAA stop codon, three (ND2, ND3, and ND6) contained TAG stop codons, and the others terminated in T − or TA − and were likely completed as TAA by post-transcriptional polyadenylation (Ojala et al. Citation1981). An 899 bp control region, or displacement loop (D-loop), was located between the tRNAPro and tRNAPhe loci. Eight of the 22 tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer2, tRNAGlu, tRNAPro) and one PCG (ND6) were encoded on the light strand. The remaining genes were encoded on the heavy strand. All tRNAs ranged from 67 to 75 bp in length, and each folded into a typical secondary structure. Two of them (tRNAMet and tRNATrp) overlapped on a single nucleotide. Sequences were clustered () with the MEGA software version 7.0.14 (Kumar et al. Citation2016) for analysis of similarity of the mitogenomes. Together they formed a sister group of H. incognitum, thus maintaining the grouping of the Family Loricariidae and the Order Siluriformes. Therefore, taxonomic and molecular investigations are crucial for its conservation. Characterization of the mtDNA genome of Hypostomus species can significantly contribute to elucidate the taxonomy of the group since only one species of the genus – H. plecostomus (Liu et al., Citation2014) have its mtDNA genome sequenced to date.
Figure 1. Tree of similarity of mitochondrial DNA (mtDNA) sequences. Comparison of the mitogenomes of ten species of the Order Siluriformes: Hypostomus Plecostomus (NC_025584.1), Hypoptopoma incognitum (NC_028072.1), Amblydoras gonzalezi (NC_015745.1), Platydoras armatulus (NC_025585.1), Centromochlus perugiae (NC_015748.1), Corydoras rabauti (NC_004698.1), Corydoras nattereri (KT239009.1), Pimelodus pictus (NC_015797.1), Pimelodus maculatus (NC_032695.1), and Lophiosilurus alexandri (KT990219.1); and five species of the Order Characiformes: Hoplias intermedius (NC_033860.1), Salminus brasiliensis (NC_024941.1), Leporinus elongatus (NC_034281.1), Brycon orgignyanus (NC_024938.1), and Prochilodus costatus (NC_027690.1). Characiform species were used as the outgroup. The consensus tree was constructed using the Kimura-2 parameter model (Kimura Citation1980) and 1000 bootstrap. The D-loop region was excluded from this analysis due to its high variability (Gonder et al. Citation2007). Hypostomus francisci grouped with H. plecostomus, and together they formed a sister group of H. incognitum, thus maintaining the Family Loricariidae as a single clade. Percentage support values for each group are indicated on the branches.
Acknowledgment
Universidade Federal de Minas Gerais (UFMG) for the support during the project execution
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Alves CBM, Pompeu PS. 2010. Peixes do Rio das Velhas: passado e presente. Belo Horizonte, Brazil: SEGRAC.
- Armbruster JW. 2004. Phylogenetic relationships of the suckermouth armoured catfishes (Loricariidae) with emphasis on the Hypostominae and the Ancistrinae. Zool J Linn Soc. 141:1–80.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Comp Sci Biol. 99:45–56.
- Dias AC, Zawadzki CH. 2018. Identification key and pictures of the Hypostomus Lacépède, 1803 (Siluriformes, Loricariidae) from the rio Ivaí, upper rio Paraná basin. CheckList. 14:393–414.
- Drummond AJB, Ashton M, Cheung J, Heled M, Kearse R, Moir S, Stones-Havas, et al. 2009. Geneious version R7.1.3 for Windows. Computer program and documentation distributed by the author. Website http://www.geneious.com [accessed 2018 Aug 29].
- Eschmeyer WN, Fricke R, van der Laan R, Fong J. 2017. Catalog of fishes: genera, species, references [Internet]. San Francisco: California Academy of Science. https://www.calacademy.org/ [accessed 2018 Sep 13].
- Gonder MK, Mortensen HM, Reed FA, Sousa A, Tishkoff SA. 2007. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 24:757–768.
- Isbrücker IJH. 2001. Nomenklator der Gattungen und Arten der Harnischwelse, Familie Loricariidae Rafinesque, 1815 (Teleostei, Ostariophysi). Die Aquarienzeitschrift (DATZ SonderheftHarnischwelse). 2:25–32.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh T, Sado T, et al. 2013. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 11:2531–2540.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu S, Zhang J, Yao J, Liu Z. 2014. The complete mitochondrial genome of the armored catfish, Hypostomus plecostomus (Siluriformes: Loricariidae). Mitochondrial DNA. 27:1–2. 10.3109/19401736.2014.971281.
- Lütken CF. 1874. Siluridae novae Brasiliae centralis a clarissimo J. Reinhardt in provincia Minas-gerais circa oppidulum Lagoa Santa, praecipue in flumine Rio das Velhas et affluentibus collectae, secundum caracteres essentiales, breviter descriptae. Overs Danske Vidensk Selsk Forhandl Kjobenhavn. 29–36.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Ramos TPA, Zawadzki CH, Ramos RTC, Britski HA. 2017. Redescription of Hypostomus johnii, a senior synonym of Hypostomus eptingi (Siluriformes: Loricariidae), Northeastern Brazil. Neotrop Ichthyol. 15:e160064.
